Last active
February 28, 2019 15:26
-
-
Save FilipDominec/c3663987f3a48325f840addfd193994c to your computer and use it in GitHub Desktop.
Loads ASCII-exported spectra from Princeton spectrometer, exports "worm plot" of spectral position and intensity of the peak
This file contains hidden or bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| #!/usr/bin/python3 | |
| #-*- coding: utf-8 -*- | |
| """ | |
| Batch fitting of multiple spectra - evaluation of spectroscopic response of InGaN/GaN quantum-well sensors to the | |
| presence of surface charges induced by gases or liquids. | |
| Invocation: | |
| ./multifit.py <filename>.dat | |
| where | |
| filename = ASCII data file to be processed, in the format exported by Princeton monochromator of four columns | |
| corresponding to: 0. wavelength, 1. numbering of spectral acquisition, 2. nothing, 3. rel. intensity | |
| Output: | |
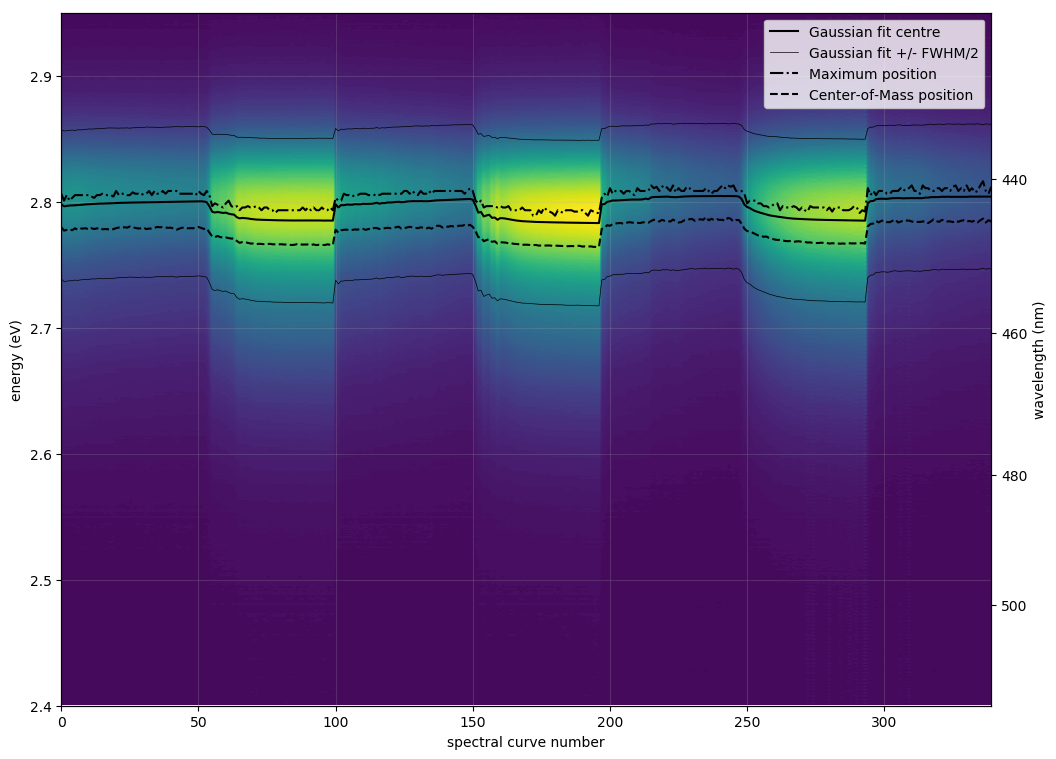
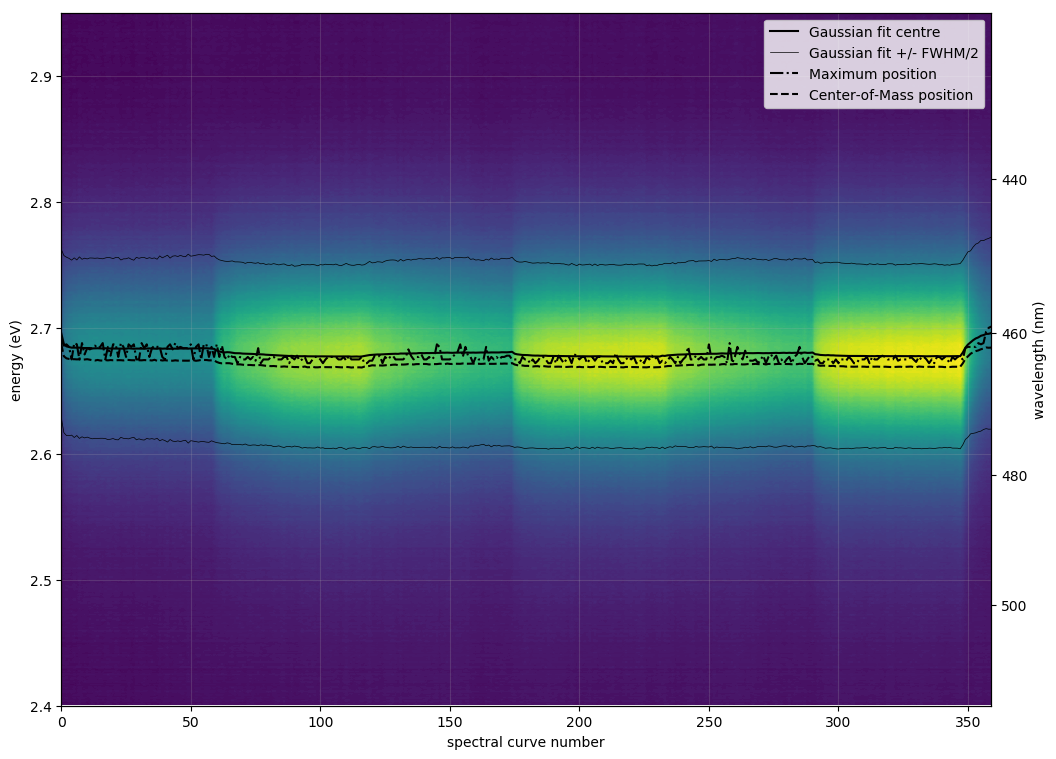
| <filename>contours.png | |
| = spectrum as background contours, detected emission energy as black line | |
| <filename>_fit.txt | |
| = fitting results: n, Ifit, E0fit, FWHMfit, Ip, Ep, Is,Ecom, | |
| n - number of spectrum | |
| Ifit, E0fit, FWHMfit - fitting results, see fitting function in the code below | |
| Ip, Ep - intensity and energy of 'peak point', i.e. absolute maximum | |
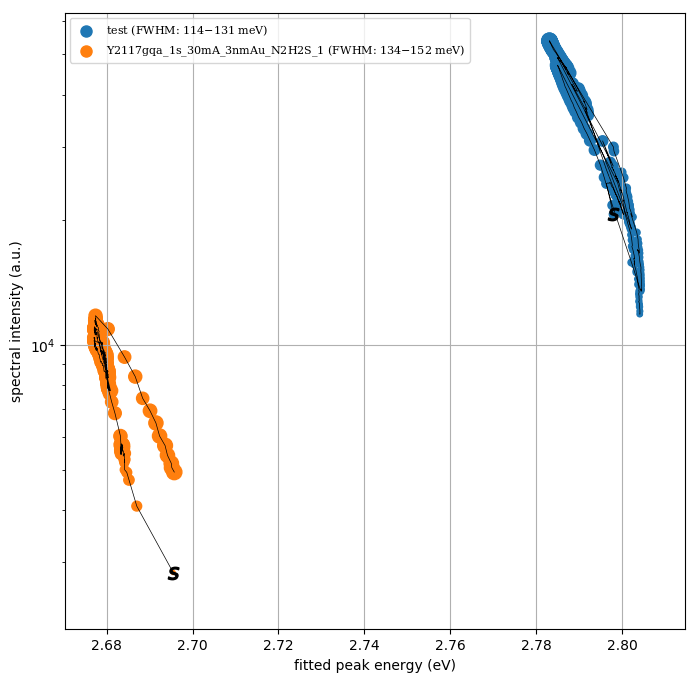
| <filename>_worms.png | |
| <filename>_worms.pdf | |
| = the worm plot Is, Ecom - summed intensity and center-of-mass position | |
| """ | |
| ## Import common moduli | |
| import matplotlib, sys, os, time | |
| from pathlib import Path | |
| import matplotlib.pyplot as plt | |
| import numpy as np | |
| from scipy.constants import c, hbar, pi | |
| ## ========== User settings =========== | |
| ## Single-Gaussian fitting, similar as above, but no phonon replicas (samples differ by their spectra from the model) | |
| ## I - peak integral intensity | |
| ## E0 - central energy | |
| ## FWHM - full width at half maximum of the major peak | |
| ## R - describe the ratio of consecutive LO-phonon replicas in GaN, each red-shifted by 0.092 eV | |
| def fit_func(E, I, E0, FWHM): | |
| return I/FWHM* np.exp(-(E-E0)**2/(FWHM/2)**2 * np.log(2)) | |
| fit_bounds = ([0, 1240/490, .001], [1e7, 1240/420, .4]) | |
| file_note = '' | |
| ## Plotting options | |
| worm_yscale = 'log' # Logarithmic intensity axis for wormplots may better enable to compare sample evolution | |
| ylim = (2.4, 2.95) # Energy limits for plotting (just visual) | |
| ## Spectral clipping to the visible region (clips actual data - affects fitting) | |
| clipcenter, clipwidth = (ylim[0]+ylim[1])/2, (ylim[1]-ylim[0]) # note: use high value for clipwidth to disable clipping | |
| ## Debugging options: | |
| limit_number_of_spectra = False # set to False to process spectra up to the end | |
| pick_each_nth_spectrum = 1 # use 1 to process each spectrum | |
| debugmode = False # Will also export a diagnostics of the selected spectral fit | |
| ## Experimental features for future | |
| flatten_rel_file_path = False | |
| subtract_background = False # experimental! | |
| ## Experimental: uncomment for double-peak fitting (may need fine-tuning to converge!) | |
| #def fit_func(E, I, E0, FWHM, I2, DeltaE, FWHM2): ## Experimental | |
| #return I /FWHM* np.exp(-(E -E0)**2/(FWHM /2)**2 * np.log(2)) + \ | |
| #I /FWHM * np.exp(-(E -E0)**2/(FWHM /2)**2 * np.log(2)) | |
| #fit_bounds = ([0, 2.7, .001, 0, 0.05, .001], [1e6, 2.9, .3, 1e6, 0.25, .2]) | |
| #file_note = 'double_gaussian' | |
| ## ========== End of user settings =========== | |
| ## Three different plotting functions | |
| def plot_diagnostic(): | |
| fig = plt.figure(); ax = plt.subplot(111) | |
| plt.plot(energy_eV, intensity, 'b-') # The measured spectrum | |
| if 'file_note' != 'double_gaussian': | |
| plt.plot(energy_eV, fit_func(energy_eV, *popt), 'g--', label='fit: I=%5.3f, E0=%5.3f, FWHM=%5.3f' % tuple(popt)) | |
| else : | |
| ax.plot(energy_eV, fit_func(energy_eV, *popt), 'k--', | |
| label='fit: I=%5.3f, E0=%5.3f, FWHM=%5.3f, I=%5.3f, E0=%5.3f, FWHM=%5.3f' % tuple(popt)) | |
| popt2 = popt.copy(); popt2[0]= 0 # First Gaussian separately | |
| ax.plot(energy_eV, fit_func(energy_eV, *popt2), 'r--', | |
| label='fit: I=%5.3f, E0=%5.3f, FWHM=%5.3f, I=%5.3f, E0=%5.3f, FWHM=%5.3f' % tuple(popt)) | |
| popt2 = popt.copy(); popt2[3]= 0 # Second Gaussian separately | |
| ax.plot(energy_eV, fit_func(energy_eV, *popt2), 'g--', | |
| label='fit: I=%5.3f, E0=%5.3f, FWHM=%5.3f, I=%5.3f, E0=%5.3f, FWHM=%5.3f' % tuple(popt)) | |
| #fig.title(popt) # todo | |
| plt.savefig(outfilename + file_note + '_fit_diagnostic{:}.png'.format(num)) | |
| #plt.show() | |
| def plot_all_spectra_2D(): | |
| fig, ax = plt.subplots(figsize=(12, 9)) | |
| ax.contourf(np.arange(n_spec)[:num+1], energy_eV, intensities[:num+1,:].T, levels=np.linspace(np.min(intensities), np.max(intensities), 100)) | |
| ax.plot(np.arange(n_spec)[:num+1:], results_for_file[:,2], ls='-', c='k', label='Gaussian fit centre') | |
| ax.plot(np.arange(n_spec)[:num+1], results_for_file[:,2]+results_for_file[:,3]/2, ls='-', c='k', lw=.5, label='Gaussian fit +/- FWHM/2') | |
| ax.plot(np.arange(n_spec)[:num+1], results_for_file[:,2]-results_for_file[:,3]/2, ls='-', c='k', lw=.5, label='') | |
| ax.plot(np.arange(n_spec)[:num+1], results_for_file[:,5], ls='-.', c='k', label='Maximum position') | |
| ax.plot(np.arange(n_spec)[:num+1], results_for_file[:,7], ls='--', c='k', label='Center-of-Mass position') | |
| if ylim: plt.ylim(ylim) # Force y-limit of plotting | |
| plt.grid(alpha=.2) | |
| plt.xlabel(u"spectral curve number"); | |
| plt.ylabel(u"energy (eV)"); | |
| plt.legend(prop={'size':10}, loc='upper right') | |
| ## This is an optional ugly hack to get a secondary y-axis for wavelength | |
| def right_tick_function(p): return 1240/p | |
| ax2 = ax.twinx() | |
| ax2.set_ylim(np.array(ax.get_ylim())) | |
| ax2.set_ylabel('wavelength (nm)') | |
| ## If we wish nice round numbers on the secondary axis ticks... | |
| right_ax_limits = sorted([right_tick_function(lim) for lim in ax.get_ylim()]) | |
| yticks2 = matplotlib.ticker.MaxNLocator(nbins=20, steps=[1,2,5]).tick_values(*right_ax_limits) | |
| ## ... we must give them correct positions. To do so, we need to numerically invert the tick values: | |
| from scipy.optimize import brentq | |
| def right_tick_function_inv(r, target_value=0): | |
| return brentq(lambda r,q: right_tick_function(r) - target_value, ax.get_ylim()[0], ax.get_ylim()[1], 1e6) | |
| valid_ytick2loc, valid_ytick2val = [], [] | |
| for ytick2 in yticks2: | |
| try: | |
| valid_ytick2loc.append(right_tick_function_inv(0, ytick2)) | |
| valid_ytick2val.append(ytick2) | |
| except ValueError: ## (skip tick if the ticker.MaxNLocator gave invalid target_value to brentq optimization) | |
| pass | |
| ax2.set_yticks(valid_ytick2loc) ## Finally, we set the positions and tick values | |
| from matplotlib.ticker import FixedFormatter | |
| ax2.yaxis.set_major_formatter(FixedFormatter(["%g" % righttick for righttick in valid_ytick2val])) | |
| figure_name = outfilename+file_note+'_contours.png' | |
| gprint(" 2D spectral data plotted to " + figure_name + '\n') | |
| fig.savefig(outfilename+file_note+'_contours.png', bbox_inches='tight') | |
| fig.savefig(outfilename+file_note+'_contours.pdf', bbox_inches='tight') | |
| def plot_worm(results_for_batch, labels): | |
| clip = 0 | |
| step = 1 | |
| fig = plt.figure(figsize=(8,8)); ax = plt.subplot(111) | |
| matplotlib.rc('font', size=12, family='serif') | |
| for results_for_file, label in zip(results_for_batch, labels): | |
| ny = results_for_file[clip::step,1] # intensity | |
| nx = results_for_file[clip::step,2] # spectral position | |
| w = results_for_file[clip::step,3] # spectral width | |
| nw = w-np.min(w)*0.99 # remember spectral width intensity | |
| ll = "%s" % (label.replace('0s','0185ms').replace('FWHM','').split('.dat')[0]) + \ | |
| (" (FWHM: {:3.0f}$-${:3.0f} meV)".format(np.min(w[clip:])*1e3,np.max(w[clip:])*1e3)) | |
| if 'NOLABEL' in ll: | |
| ll = '' | |
| else: | |
| ll = ll.rsplit('/',1)[-1].replace(' fit.txt','').replace('.txt','') | |
| ax.scatter(nx, ny, label=ll, s=120*nw/np.max(nw), marker='o', alpha=1) | |
| ax.scatter(nx[0], ny[0], label="", color='k', s=80, marker='$s$', alpha=1) | |
| ax.plot(nx, ny, label="", lw=.5, c='k') | |
| if worm_yscale == 'log': | |
| ax.set_yscale('log') | |
| else: | |
| globmin = min(np.min(yy) for yy in ys[::3]) | |
| globmax = max(np.max(yy) for yy in ys[::3]) | |
| if globmin < 0.5*globmax: ax.set_ylim(ymin=0) | |
| ax.set_xlabel("fitted peak energy (eV)") | |
| ax.set_ylabel("spectral intensity (a.u.)") | |
| #plot_title = sharedlabels[-6:] ## last few labels that are shared among all curves make a perfect title | |
| ax.set_title('', fontsize=10) | |
| ax.legend(loc='best', prop={'size':8}) | |
| ax.grid(True) | |
| plt.savefig(outfilename + file_note + '_worms.png', bbox_inches='tight') # todo switch from plt. to fig. | |
| plt.savefig(outfilename + file_note + '_worms.pdf', bbox_inches='tight') | |
| ## User interaction | |
| if len(sys.argv) == 1: # if no parameters given, enter the interactive GUI mode | |
| from tkinter import filedialog | |
| from tkinter import * | |
| root = Tk() | |
| root.title('multifit.py processing report') | |
| T = Text(root, height=20, width=150) | |
| T.pack() | |
| filenames = filedialog.askopenfilenames(initialdir = os.getcwd(), | |
| title = "Select spectral data file to be processed, please", | |
| filetypes = (("DAT files","*.dat"),("all files","*.*"))) | |
| else: | |
| T = None | |
| filenames = sys.argv[1:] | |
| def gprint(*args, **kwargs): # printing in the stdout and possibly into the window | |
| if T: | |
| T.insert(END, ' '.join(args)+'\n') | |
| root.update() | |
| print(*args, **kwargs) | |
| ## Load and delinearize the spectral data (according to spectrum numbering in the 3rd column of the DAT file) | |
| results_for_batch, names_for_batch = [], [] | |
| for filename in filenames: | |
| gprint("Processing", filename) | |
| outfilename = filename.replace('.dat', '').replace('%', 'pct') | |
| if flatten_rel_file_path: ## Experimental: will put all output into the working directory | |
| outfilename = outfilename.replace('./','').replace('/','-') | |
| wl, spec_id, intens = np.genfromtxt(filename, usecols=[0,2,3], unpack=True) | |
| n_spec = int(np.max(spec_id)) | |
| energy_eV = 1239.8 / wl[:int(len(wl)/n_spec)] # nm -> eV | |
| intensities = np.reshape(intens, (n_spec,-1)) | |
| if subtract_background: | |
| bg = np.mean(intensities[:,:-10],axis=1) | |
| intensities -= np.outer(bg, np.ones_like(intensities[0])) | |
| ## If specified, apply data clipping | |
| clipwindow = np.abs(energy_eV-clipcenter) < clipwidth/2 | |
| energy_eV, intensities = energy_eV[clipwindow], intensities[:, clipwindow] | |
| ## Main cycle | |
| from scipy.optimize import curve_fit | |
| results_for_file = [] | |
| for num, intensity in enumerate(intensities): | |
| intensity = intensity*energy_eV**2 | |
| if num%pick_each_nth_spectrum != 0: continue | |
| ## (not used) peak finding | |
| E_ordered_by_I = energy_eV[np.argsort(intensity)] | |
| peak_I = np.max(intensity) | |
| peak_eV = E_ordered_by_I[-1] | |
| percentile10 = E_ordered_by_I[len(E_ordered_by_I)//10] | |
| percentile0 = E_ordered_by_I[0] | |
| ## (not used) center of mass computation | |
| sum_intensity = np.sum(intensity) | |
| center_of_mass_eV = np.sum(energy_eV*intensity)/sum_intensity | |
| ## fitting | |
| try: | |
| popt, pcov = curve_fit(fit_func, energy_eV, intensity, bounds=(fit_bounds)) | |
| if debugmode and num in list_of_explicit_fit_plots: | |
| plot_diagnostic() | |
| except Exception as e: | |
| popt = [np.NaN]*3 | |
| gprint('line %d: fit failed' % num, e) | |
| ## Record results | |
| if np.nan not in popt: | |
| results_for_spectrum = [num] + list(popt) + [peak_I, peak_eV, sum_intensity/len(intensity), center_of_mass_eV] | |
| results_for_file.append(results_for_spectrum) | |
| if limit_number_of_spectra and num > limit_number_of_spectra: break | |
| results_for_file = np.array(results_for_file) | |
| ## Outputting results | |
| names_for_batch.append(outfilename+file_note) | |
| results_for_batch.append(results_for_file) | |
| fit_name = outfilename+file_note+'_fit.txt' | |
| np.savetxt(fit_name, results_for_file, fmt="%.8g", header='#n\tI_fit\tE0_fit\tFWHM_fit\tI_peak\tE0_peak\tI_sum\tE0_center_of_mass') | |
| gprint(" fit successful, results exported to " + fit_name + '\n') | |
| plot_all_spectra_2D() | |
| plot_worm(results_for_batch, names_for_batch) | |
| gprint("Worm plot of all data exported to " + outfilename + file_note + '_worms.png' + '\n') | |
| if T: | |
| gprint("Fitting and plotting finished, this window will automatically close after 15 s.") | |
| time.sleep(15) | |
| ## Simple axes | |
| #plt.yscale('log') | |
| #plt.xlim((-0.1,1.1)); plt.xscale('linear') | |
Author
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment
Example of worm plot:
Examples of contour plots: