Relevant paper: https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0218738
Confusion about the plots showing m_inf for SHL-1, see top left
plot, red curve:
https://journals.plos.org/plosone/article/file?type=supplementary&id=info:doi/10.1371/journal.pone.0218738.s003
Appendix with equations: https://journals.plos.org/plosone/article/file?type=supplementary&id=info:doi/10.1371/journal.pone.0218738.s001
Appendix with parameters used: https://journals.plos.org/plosone/article/file?type=supplementary&id=info:doi/10.1371/journal.pone.0218738.s002
According to this information the following equation is being used:
\begin{equation*}
m_{\text{SHL1,}\infty}(V) = \frac{1}{1 - \exp{\frac{-(V - V_h}{k_a}}}
\end{equation*}but in the paper this is referred to as a Boltzmann like distribution, e.g. p. 5 above equation (4).
This function however is essentially:
\begin{equation*}
f(x) = \frac{1}{1 - \exp{\frac{-x}}}
\end{equation*}which has a singularity at
https://www.wolframalpha.com/input/?i=1+%2F+%281+-+e%5E%28-x%29%29
Which means it can’t be the function mentioned in the table / paper.
Boltzmann functions (not to be confused with Boltzmann distributions or the Maxwell-Boltzmann distribution) apparently are sigmoid like functions, see e.g.: https://www.graphpad.com/guides/prism/7/curve-fitting/reg_classic_boltzmann.htm
The function should thus be:
\begin{equation*}
m_{\text{SHL1,}\infty}(V) = \frac{1}{1 + \exp{\frac{-(V - V_h}{k_a}}}
\end{equation*}Note the + in the denominator instead of -.
Assuming this is a typo in the paper, let’s try to estimate whether the given parameters:
$V_h = 11.2$ $k_a = 14.1$
can match the plot mentioned above.
To do that we’ll extract some data points in a crude way from the plot above. Extract a cropped plot from the plot above, results in:
Now we write a small script to extract data points from this:
import flippy, os, sequtils, ggplotnim, cligen, strformat
proc toGrey(img: Image): seq[uint8] =
case img.channels
of 1: result = img.data
else:
result = newSeq[uint8](img.width * img.height)
let nChannel = img.channels
var val: int = 0
var idx = 0
for i in countup(0, img.data.high, nChannel):
# calc mean of colors
val = 0
for j in 0 ..< nChannel:
val += img.data[i + j].int
result[idx] = 255 - (val div nChannel).uint8
inc idx
proc allFalse(t: Tensor[bool]): bool =
result = true
for x in t:
if x:
return false
proc extractData(fname: string): seq[float] =
let img = loadImage(fname)
let rows = img.height
let cols = img.width
let imgTensor = toGrey(img).toTensor.reshape(rows, cols)
let colIdx = arange(0, rows)
result = newSeq[float](cols)
var i = 0
for idx, cl in enumerateAxis(imgTensor, 1):
let sel = (cl >. 0).squeeze
if not allFalse(sel):
let idxs = colIdx[sel]
let ind = mean(idxs)
result[i] = (rows - ind).float / rows.float
else:
result[i] = 0.0
inc i
proc scaleBy(s: var seq[float], low, high: float) =
discard
proc main(fname: string,
yLow = 0.0,
yHigh = 1.0,
xLow = 0.0,
xHigh = 1.0) =
var yVals = extractData(fname)
yVals.scaleBy(yLow, yHigh)
let xVals = linspace(xLow, xHigh, yVals.len)
let df = seqsToDf(xVals, yVals)
ggplot(df, aes("xVals", "yVals")) +
geom_line() +
ggsave("/tmp/extracted.pdf")
df.write_csv(&"extracted_from_{fname.extractFilename}.csv")
when isMainModule:
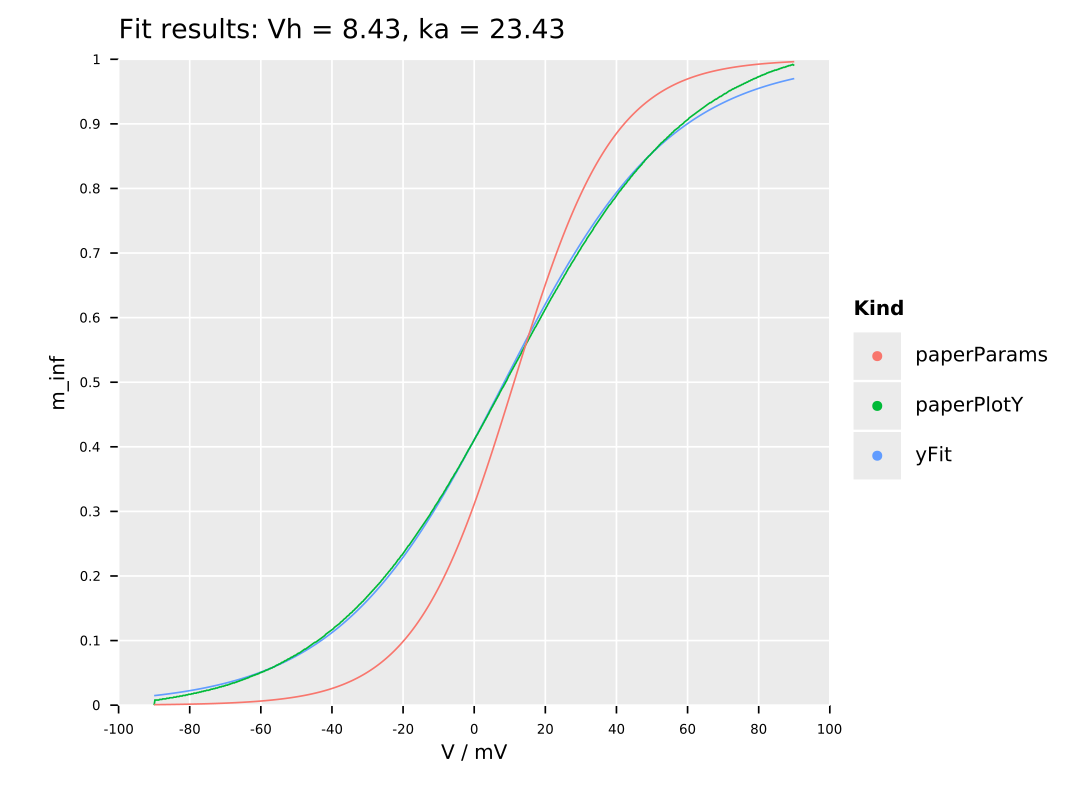
dispatch mainFinally, we can just perform a fit to those data points:
import math, ggplotnim, sequtils, strformat, mpfit
const ka = 14.1 # Parameter
const vh = 11.2
proc m_inf(V, vh, ka: float): float =
result = 1.0 / (1 + exp( -(V - vh) / ka) )
proc buildDf(fn: proc(V, vh, ka: float): float): DataFrame =
result = newDataFrame()
let xt = linspace(-90.0, 90.0, 1000)
result["xVals"] = xt
result["yVals"] = xt.map_inline(fn(x, vh, ka))
proc fitToPaperPlot() =
var df = toDf(readCsv("./extracted_from_m_inf_extract.png.csv"))
# wrap the m_inf proc to fit
let fnToFit = proc(p_ar: seq[float], x: float): float =
result = m_inf(V = x, vh = p_ar[0], ka = p_ar[1])
let xVals = df["xVals"].toTensor(float)
let yVals = df["paperPlotY"].toTensor(float)
let yErr = yVals.map_inline(1e-3) # guesstimate error...
# fit to the data, start with paper parameters
let (pRes, res) = fit(fnToFit,
@[11.2, 14.1],
xVals.toRawSeq,
yVals.toRawSeq,
yErr.toRawSeq)
df["yFit"] = xVals.map_inline(fnToFit(pRes, x))
df = df.gather(["paperPlotY", "yFit"], key = "Kind", value = "yVals")
# add paper values
var dfPaper = buildDf(m_inf)
dfPaper["Kind"] = constantColumn("paperParams", dfPaper.len)
echo dfPaper
echo df
df.add dfPaper
echoResult(pRes, res = res)
ggplot(df, aes("xVals", "yVals", color = "Kind")) +
geom_line() +
ylab("m_inf") +
xlab("V / mV") +
ggtitle(&"Fit results: Vh = {pRes[0]:.2f}, ka = {pRes[1]:.2f}") +
theme_opaque() +
ggsave("/tmp/neuron_fit.png")
when isMainModule:
fitToPaperPlot()Which gives us the following final result: