Last active
April 27, 2023 12:55
-
-
Save ahwillia/d3e13896f990e9676cad77ab260295d8 to your computer and use it in GitHub Desktop.
PCA and NMF with cross-validation (using Numpy)
This file contains hidden or bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| from pca_crossval import * | |
| from tqdm import tqdm | |
| import itertools | |
| np.random.seed(1111) | |
| m, n, r = 100, 101, 3 | |
| Utrue, Vtrue = rand(m,r), rand(r,n) | |
| data = np.dot(Utrue, Vtrue) + .25*randn(m,n) | |
| data[data < 0] = 0 | |
| ranks = [] | |
| train_err = [] | |
| test_err = [] | |
| rank_range = range(1,11) | |
| repeat_range = range(10) | |
| with tqdm(total=len(rank_range)*len(repeat_range)) as pbar: | |
| for rank, repeat in itertools.product(rank_range, repeat_range): | |
| _, _, train, test = crossval_pca(data, rank, nonneg=True) | |
| ranks.append(rank) | |
| train_err.append(train[-1]) | |
| test_err.append(test[-1]) | |
| pbar.update(1) | |
| ranks = np.array(ranks) | |
| train_err = np.array(train_err) | |
| test_err = np.array(test_err) | |
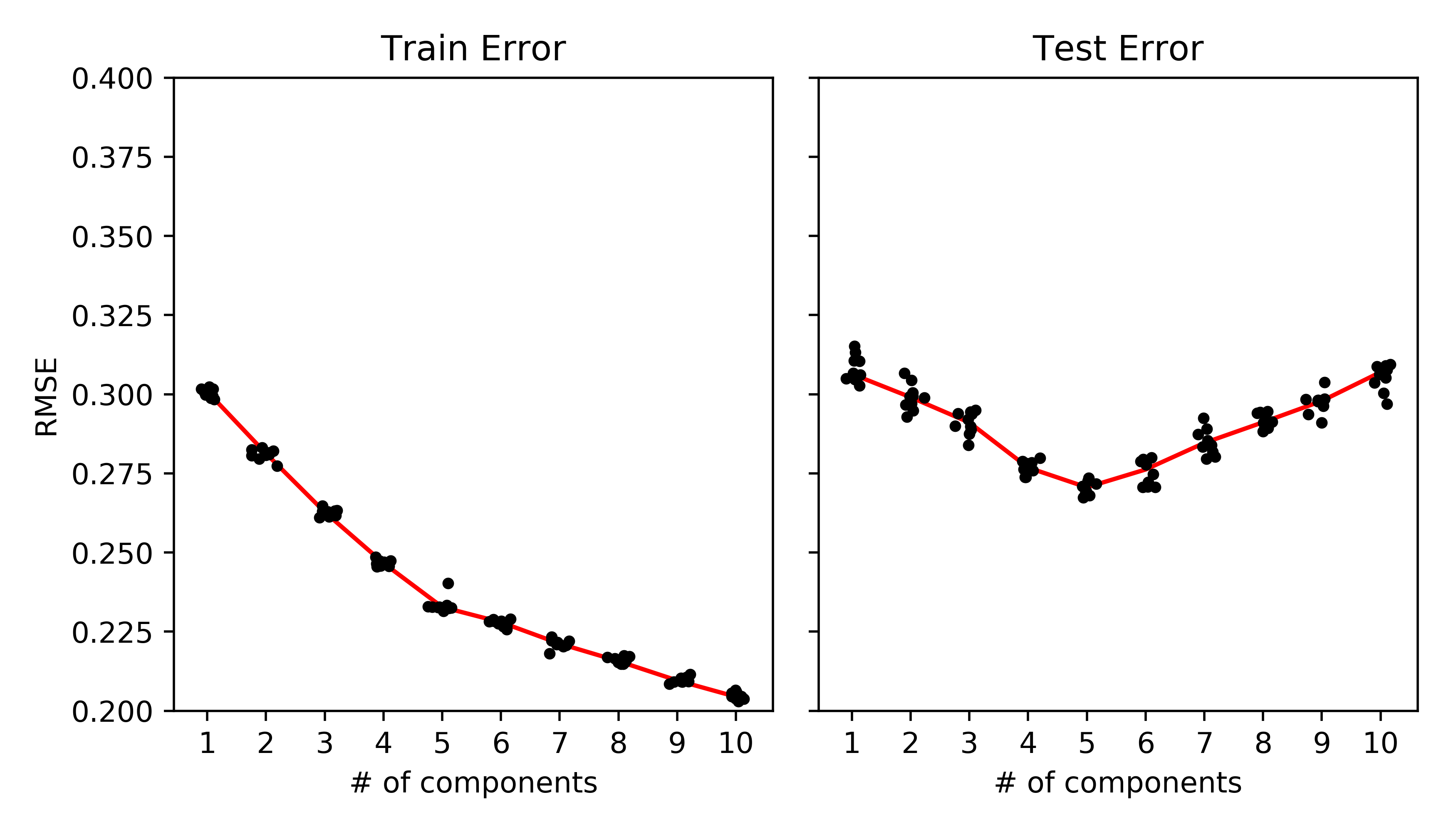
| fig, axes = plt.subplots(1, 2, sharey=True, sharex=True, figsize=(7, 4)) | |
| axes[0].plot(ranks + randn(ranks.size)*.1, train_err, '.k') | |
| axes[0].plot(np.unique(ranks), [np.mean(train_err[ranks==r]) for r in np.unique(ranks)], '-r', zorder=-1) | |
| axes[0].set_ylabel('RMSE') | |
| axes[0].set_title('Train Error') | |
| axes[0].set_xlabel('# of components') | |
| axes[1].plot(ranks + randn(ranks.size)*.1, test_err, '.k') | |
| axes[1].plot(np.unique(ranks), [np.mean(test_err[ranks==r]) for r in np.unique(ranks)], '-r', zorder=-1) | |
| axes[1].set_xticks(np.unique(ranks).astype(int)) | |
| axes[1].set_title('Test Error') | |
| axes[1].set_xlabel('# of components') | |
| fig.tight_layout() | |
| fig.savefig('cv_nmf.pdf') | |
| plt.show() |
This file contains hidden or bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| from pca_crossval import * | |
| import itertools | |
| from tqdm import tqdm | |
| np.random.seed(2222) | |
| m, n, r = 500, 501, 5 | |
| Utrue, Vtrue = randn(m,r), randn(r,n) | |
| data = np.dot(Utrue, Vtrue) + 6*randn(m,n) | |
| ranks = [] | |
| train_err = [] | |
| test_err = [] | |
| rank_range = range(1,11) | |
| repeat_range = range(10) | |
| with tqdm(total=len(rank_range)*len(repeat_range)) as pbar: | |
| for rank, repeat in itertools.product(rank_range, repeat_range): | |
| _, _, train, test = crossval_pca(data, rank) | |
| ranks.append(rank) | |
| train_err.append(train[-1]) | |
| test_err.append(test[-1]) | |
| pbar.update(1) | |
| ranks = np.array(ranks) | |
| train_err = np.array(train_err) | |
| test_err = np.array(test_err) | |
| fig, axes = plt.subplots(1, 2, sharey=True, sharex=True, figsize=(7, 4)) | |
| axes[0].plot(ranks + randn(ranks.size)*.1, train_err, '.k') | |
| axes[0].plot(np.unique(ranks), [np.mean(train_err[ranks==r]) for r in np.unique(ranks)], '-r', zorder=-1) | |
| axes[0].set_ylabel('RMSE') | |
| axes[0].set_title('Train Error') | |
| axes[0].set_xlabel('# of components') | |
| axes[1].plot(ranks + randn(ranks.size)*.1, test_err, '.k') | |
| axes[1].plot(np.unique(ranks), [np.mean(test_err[ranks==r]) for r in np.unique(ranks)], '-r', zorder=-1) | |
| axes[1].set_xticks(np.unique(ranks).astype(int)) | |
| axes[1].set_title('Test Error') | |
| axes[1].set_xlabel('# of components') | |
| fig.tight_layout() | |
| fig.savefig('cv_pca.pdf') | |
| plt.show() |
This file contains hidden or bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| import numpy as np | |
| from numpy.random import randn, rand | |
| from scipy.optimize import minimize | |
| import matplotlib.pyplot as plt | |
| def censored_least_squares(A, X0, B, M, options=dict(maxiter=20), **kwargs): | |
| """Approximately solves least-squares with missing/censored data by L-BFGS-B | |
| Updates X to minimize Frobenius norm of M .* (A*X - B), where | |
| M is a masking matrix (m x n filled with zeros and ones), A and | |
| B are constant matrices. | |
| Parameters | |
| ---------- | |
| A : ndarray | |
| m x k matrix | |
| X0 : ndarray | |
| k x n matrix, initial guess for X | |
| B : ndarray | |
| m x n matrix | |
| M : ndarray | |
| m x n matrix, filled with zeros and ones | |
| options : dict | |
| optimization options passed to scipy.optimize.minimize | |
| Note: additional keyword arguments are passed to scipy.optimize.minimize | |
| Returns | |
| ------- | |
| result : OptimizeResult | |
| returned by scipy.optimize.minimize | |
| """ | |
| k = A.shape[1] | |
| n = B.shape[1] | |
| def fg(x): | |
| X = x.reshape(k, n) | |
| resid = np.dot(A, X) - B | |
| f = 0.5*np.sum(resid[M]**2) | |
| g = np.dot(A.T, (M * resid)) | |
| return f, g.ravel() | |
| return minimize(fg, X0.ravel(), method='L-BFGS-B', jac=True, options=options, **kwargs) | |
| def crossval_pca(data, rank, p_holdout=.25, tol=1e-4, nonneg=False): | |
| """Fits PCA or NMF while holding out data at random | |
| Finds U and Vt that minimize Frobenius norm of (U*Vt - data) over | |
| a random subset of the entries in data. | |
| Note: this code does not return orthogonalized factors for PCA. | |
| Parameters | |
| ---------- | |
| data : ndarray | |
| m x n matrix | |
| rank : int | |
| number of components | |
| p_holdout : float | |
| probability of holding out an entry, expected proportion of data in test set. | |
| tol: float | |
| absolute convergence criterion on the root-mean-square-error on training set | |
| nonneg : bool | |
| if True, fit NMF model | |
| Returns | |
| ------- | |
| U : ndarray | |
| m x rank matrix | |
| Vt : ndarray | |
| rank x n matrix | |
| train_hist : list | |
| RMSE on training set on each iteration | |
| test_hist : list | |
| RMSE on test set on each iteration | |
| """ | |
| # m = num observations, n = num features | |
| m, n = data.shape | |
| # initialize factor matrices | |
| U, Vt = randn(m, rank), randn(rank, n) | |
| # hold out data at random | |
| M = rand(m, n) > p_holdout | |
| # initial loss | |
| converged = False | |
| resid = np.dot(U, Vt) - data | |
| train_hist = [np.sqrt(np.mean((resid[M])**2))] | |
| test_hist = [np.sqrt(np.mean((resid[~M])**2))] | |
| # impose nonnegativity, if desired | |
| if nonneg: | |
| bounds_U = [(0, None) for _ in range(U.size)] | |
| bounds_V = [(0, None) for _ in range(Vt.size)] | |
| else: | |
| bounds_U = None | |
| bounds_V = None | |
| # optimize | |
| while not converged: | |
| # update V | |
| r = censored_least_squares(U, Vt, data, M, bounds=bounds_V) | |
| Vt = r.x.reshape(rank, n) | |
| # update U | |
| r = censored_least_squares(Vt.T, U.T, data.T, M.T, bounds=bounds_U) | |
| U = r.x.reshape(rank, m).T | |
| # recprd train/test error | |
| resid = np.dot(U, Vt) - data | |
| train_hist.append(np.sqrt(np.mean((resid[M])**2))) | |
| test_hist.append(np.sqrt(np.mean((resid[~M])**2))) | |
| converged = (train_hist[-2]-train_hist[-1]) < tol | |
| return U, Vt, train_hist, test_hist |
Author
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment
PCA example. (Takes ~3 mins on my laptop for 500x501 rank-5 matrix)
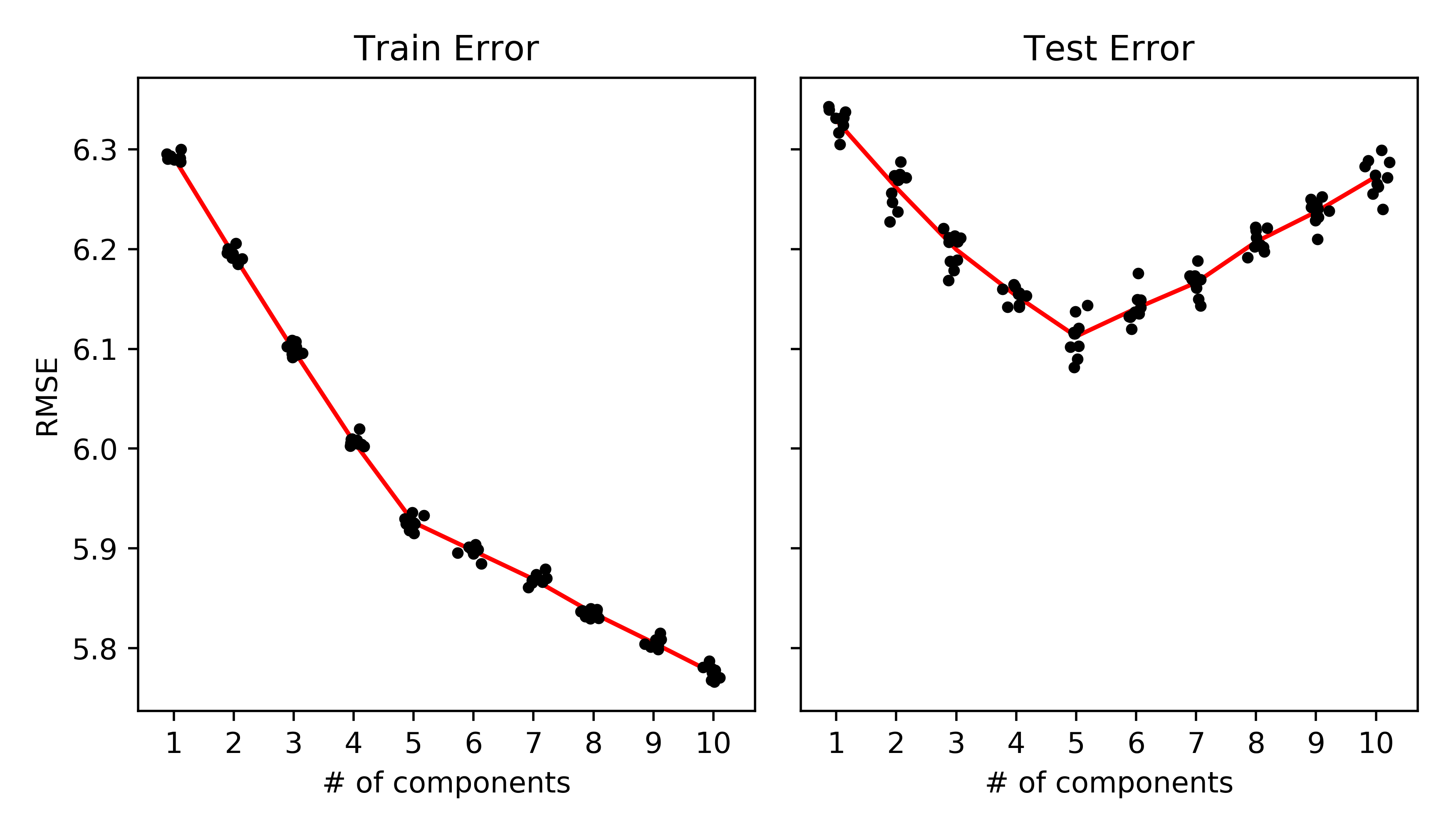
NMF example. (Takes ~30 s on my laptop for 100x101 rank-5 matrix)