library("scRNAseq")
#> Loading required package: SingleCellExperiment
#> Loading required package: SummarizedExperiment
#> Loading required package: MatrixGenerics
#> Loading required package: matrixStats
#>
#> Attaching package: 'MatrixGenerics'
#> The following objects are masked from 'package:matrixStats':
#>
#> colAlls, colAnyNAs, colAnys, colAvgsPerRowSet, colCollapse,
#> colCounts, colCummaxs, colCummins, colCumprods, colCumsums,
#> colDiffs, colIQRDiffs, colIQRs, colLogSumExps, colMadDiffs,
#> colMads, colMaxs, colMeans2, colMedians, colMins, colOrderStats,
#> colProds, colQuantiles, colRanges, colRanks, colSdDiffs, colSds,
#> colSums2, colTabulates, colVarDiffs, colVars, colWeightedMads,
#> colWeightedMeans, colWeightedMedians, colWeightedSds,
#> colWeightedVars, rowAlls, rowAnyNAs, rowAnys, rowAvgsPerColSet,
#> rowCollapse, rowCounts, rowCummaxs, rowCummins, rowCumprods,
#> rowCumsums, rowDiffs, rowIQRDiffs, rowIQRs, rowLogSumExps,
#> rowMadDiffs, rowMads, rowMaxs, rowMeans2, rowMedians, rowMins,
#> rowOrderStats, rowProds, rowQuantiles, rowRanges, rowRanks,
#> rowSdDiffs, rowSds, rowSums2, rowTabulates, rowVarDiffs, rowVars,
#> rowWeightedMads, rowWeightedMeans, rowWeightedMedians,
#> rowWeightedSds, rowWeightedVars
#> Loading required package: GenomicRanges
#> Loading required package: stats4
#> Loading required package: BiocGenerics
#>
#> Attaching package: 'BiocGenerics'
#> The following objects are masked from 'package:stats':
#>
#> IQR, mad, sd, var, xtabs
#> The following objects are masked from 'package:base':
#>
#> anyDuplicated, append, as.data.frame, basename, cbind, colnames,
#> dirname, do.call, duplicated, eval, evalq, Filter, Find, get, grep,
#> grepl, intersect, is.unsorted, lapply, Map, mapply, match, mget,
#> order, paste, pmax, pmax.int, pmin, pmin.int, Position, rank,
#> rbind, Reduce, rownames, sapply, setdiff, sort, table, tapply,
#> union, unique, unsplit, which.max, which.min
#> Loading required package: S4Vectors
#>
#> Attaching package: 'S4Vectors'
#> The following objects are masked from 'package:base':
#>
#> expand.grid, I, unname
#> Loading required package: IRanges
#> Loading required package: GenomeInfoDb
#> Loading required package: Biobase
#> Welcome to Bioconductor
#>
#> Vignettes contain introductory material; view with
#> 'browseVignettes()'. To cite Bioconductor, see
#> 'citation("Biobase")', and for packages 'citation("pkgname")'.
#>
#> Attaching package: 'Biobase'
#> The following object is masked from 'package:MatrixGenerics':
#>
#> rowMedians
#> The following objects are masked from 'package:matrixStats':
#>
#> anyMissing, rowMedians
library("scater")
#> Loading required package: scuttle
#> Loading required package: ggplot2
library("scran")
library("flexmix")
#> Loading required package: lattice
library("mixtools")
#> mixtools package, version 1.2.0, Released 2020-02-05
#> This package is based upon work supported by the National Science Foundation under Grant No. SES-0518772.
library("viridis")
#> Loading required package: viridisLite
library("annotables")
#> Error in library("annotables"): there is no package called 'annotables'
theme_set(theme_bw())
sce <- ZeiselBrainData()
#> snapshotDate(): 2022-04-26
#> see ?scRNAseq and browseVignettes('scRNAseq') for documentation
#> loading from cache
#> see ?scRNAseq and browseVignettes('scRNAseq') for documentation
#> loading from cache
#> see ?scRNAseq and browseVignettes('scRNAseq') for documentation
#> loading from cache
#> snapshotDate(): 2022-04-26
#> see ?scRNAseq and browseVignettes('scRNAseq') for documentation
#> loading from cache
sce <- addPerCellQC(sce, subsets = list(mito = grepl("^mt-", rownames(sce))))
plot(sce$detected, sce$subsets_mito_percent)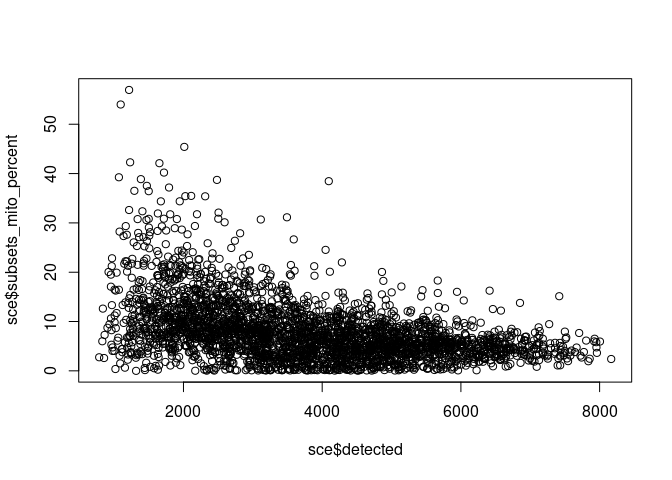
mix <- flexmix(subsets_mito_percent ~ detected, data = colData(sce), k = 2)
plot(mix)
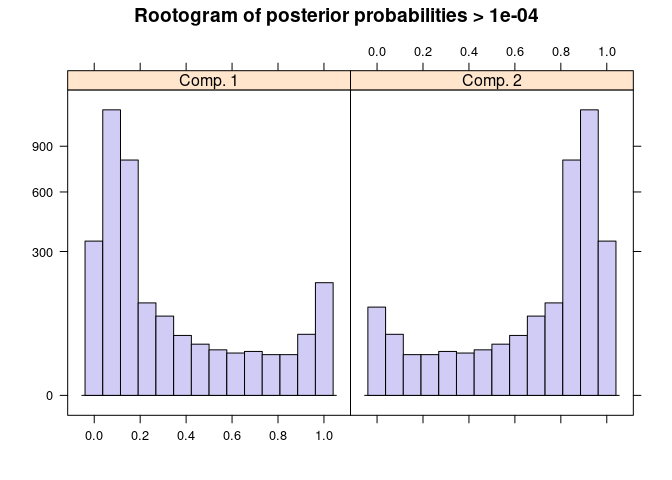
c1 <- parameters(mix, component = 1)
c2 <- parameters(mix, component = 2)
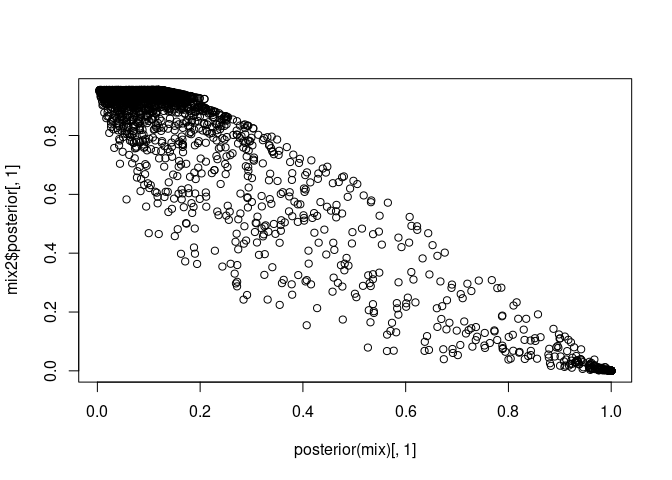
mix2 <- mixtools::normalmixEM(sce$subsets_mito_percent)
#> number of iterations= 76
plot(posterior(mix)[, 1], mix2$posterior[, 1])
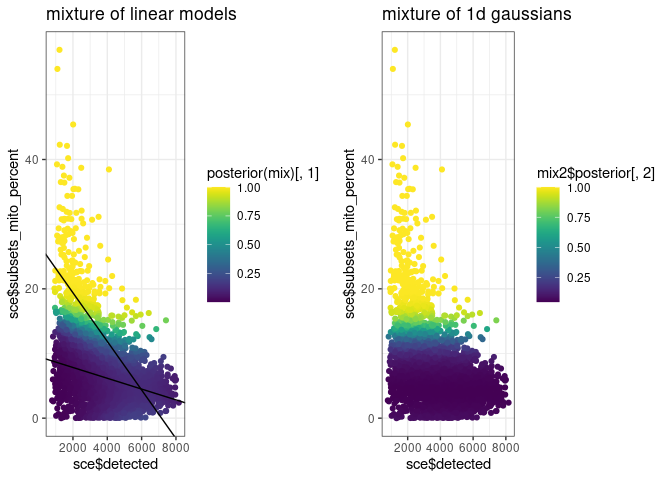
g1 <- ggplot() +
geom_point(aes(sce$detected, sce$subsets_mito_percent, colour = posterior(mix)[, 1])) +
geom_abline(intercept = c1[[1]], slope = c1[[2]]) +
geom_abline(intercept = c2[[1]], slope = c2[[2]]) +
scale_colour_viridis()
g2 <- ggplot() +
geom_point(aes(sce$detected, sce$subsets_mito_percent, colour = mix2$posterior[, 2])) +
scale_colour_viridis()
post_diff <- posterior(mix)[, 1] - mix2$posterior[, 2]
ma <- max(abs(post_diff))
ggplot() +
geom_point(aes(sce$detected, sce$subsets_mito_percent, colour = post_diff)) +
scale_colour_distiller(palette = "RdYlBu", limits = c(-ma, ma))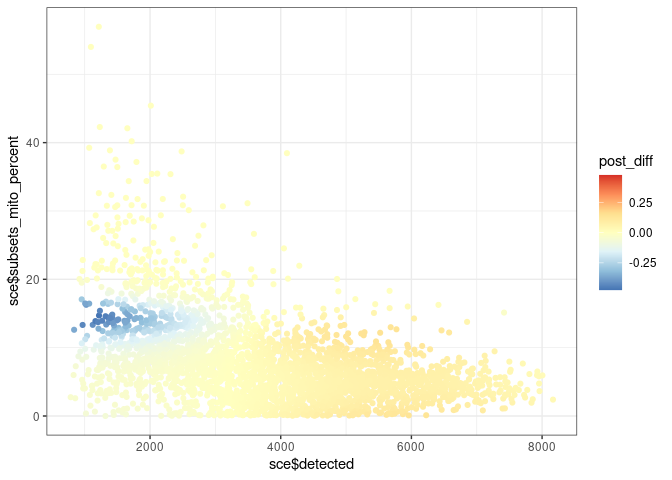
cowplot::plot_grid(
g1 + ggtitle("mixture of linear models"),
g2 + ggtitle("mixture of 1d gaussians")
)
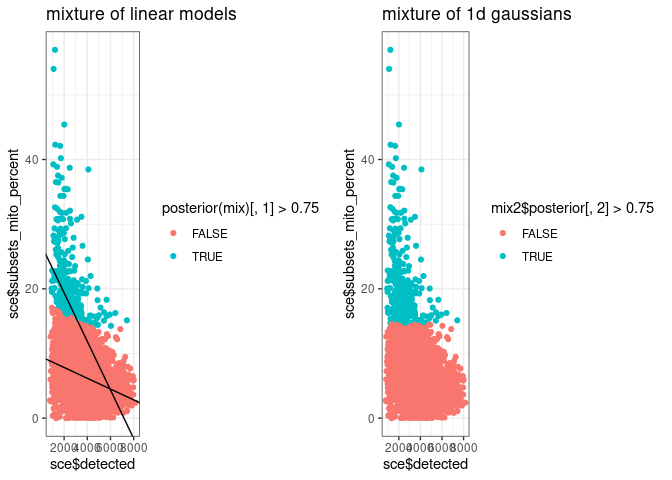
g3 <- ggplot() +
geom_point(aes(sce$detected, sce$subsets_mito_percent, colour = posterior(mix)[, 1] > 0.75)) +
geom_abline(intercept = c1[[1]], slope = c1[[2]]) +
geom_abline(intercept = c2[[1]], slope = c2[[2]])
g4 <- ggplot() +
geom_point(aes(sce$detected, sce$subsets_mito_percent, colour = mix2$posterior[, 2] > 0.75))
cowplot::plot_grid(
g3 + ggtitle("mixture of linear models"),
g4 + ggtitle("mixture of 1d gaussians")
)
mix2d <- mvnormalmixEM(colData(sce)[, c("subsets_mito_percent", "detected")], k = 2)
#> number of iterations= 94
plot(mix2d, whichplots=2)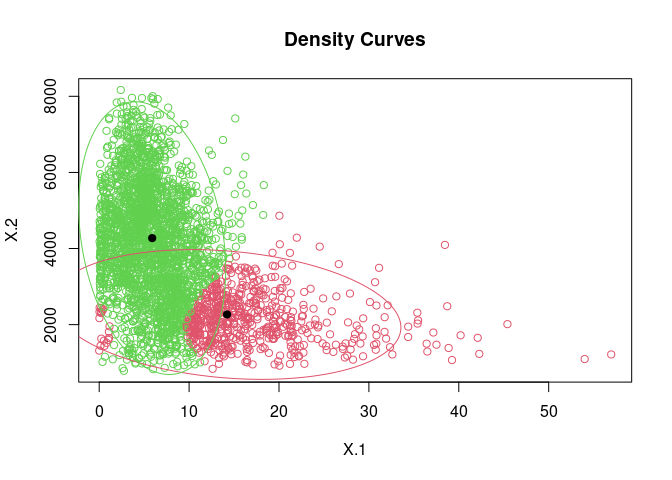
mix2dnp <- mvnpEM(colData(sce)[, c("subsets_mito_percent", "detected")], mu0 = 2)
#> iteration 1 : lambda 0.5594 0.4406
#> iteration 2 : lambda 0.5544 0.4456
#> iteration 3 : lambda 0.5474 0.4526
#> iteration 4 : lambda 0.5389 0.4611
#> iteration 5 : lambda 0.5295 0.4705
#> iteration 6 : lambda 0.5196 0.4804
#> iteration 7 : lambda 0.5098 0.4902
#> iteration 8 : lambda 0.5002 0.4998
#> iteration 9 : lambda 0.4911 0.5089
#> iteration 10 : lambda 0.4828 0.5172
#> iteration 11 : lambda 0.4751 0.5249
#> iteration 12 : lambda 0.468 0.532
#> iteration 13 : lambda 0.4616 0.5384
#> iteration 14 : lambda 0.4558 0.5442
#> iteration 15 : lambda 0.4505 0.5495
#> iteration 16 : lambda 0.4456 0.5544
#> iteration 17 : lambda 0.441 0.559
#> iteration 18 : lambda 0.4369 0.5631
#> iteration 19 : lambda 0.433 0.567
#> iteration 20 : lambda 0.4294 0.5706
#> iteration 21 : lambda 0.4261 0.5739
#> iteration 22 : lambda 0.4229 0.5771
#> iteration 23 : lambda 0.42 0.58
#> iteration 24 : lambda 0.4172 0.5828
#> iteration 25 : lambda 0.4146 0.5854
#> iteration 26 : lambda 0.4122 0.5878
#> iteration 27 : lambda 0.4099 0.5901
#> iteration 28 : lambda 0.4078 0.5922
#> iteration 29 : lambda 0.4057 0.5943
#> iteration 30 : lambda 0.4038 0.5962
#> iteration 31 : lambda 0.402 0.598
#> iteration 32 : lambda 0.4003 0.5997
#> iteration 33 : lambda 0.3987 0.6013
#> iteration 34 : lambda 0.3972 0.6028
#> iteration 35 : lambda 0.3958 0.6042
#> iteration 36 : lambda 0.3944 0.6056
#> iteration 37 : lambda 0.3932 0.6068
#> iteration 38 : lambda 0.392 0.608
#> iteration 39 : lambda 0.3909 0.6091
#> iteration 40 : lambda 0.3898 0.6102
#> iteration 41 : lambda 0.3888 0.6112
#> iteration 42 : lambda 0.3878 0.6122
#> iteration 43 : lambda 0.387 0.613
#> iteration 44 : lambda 0.3861 0.6139
#> iteration 45 : lambda 0.3853 0.6147
#> iteration 46 : lambda 0.3846 0.6154
#> iteration 47 : lambda 0.3839 0.6161
#> iteration 48 : lambda 0.3832 0.6168
#> iteration 49 : lambda 0.3826 0.6174
#> iteration 50 : lambda 0.382 0.618
#> iteration 51 : lambda 0.3814 0.6186
#> iteration 52 : lambda 0.3809 0.6191
#> iteration 53 : lambda 0.3804 0.6196
#> iteration 54 : lambda 0.3799 0.6201
#> iteration 55 : lambda 0.3795 0.6205
#> iteration 56 : lambda 0.379 0.621
#> iteration 57 : lambda 0.3786 0.6214
#> iteration 58 : lambda 0.3783 0.6217
#> iteration 59 : lambda 0.3779 0.6221
#> iteration 60 : lambda 0.3776 0.6224
#> iteration 61 : lambda 0.3773 0.6227
#> iteration 62 : lambda 0.377 0.623
#> iteration 63 : lambda 0.3767 0.6233
#> iteration 64 : lambda 0.3764 0.6236
#> iteration 65 : lambda 0.3762 0.6238
#> iteration 66 : lambda 0.3759 0.6241
#> iteration 67 : lambda 0.3757 0.6243
#> iteration 68 : lambda 0.3755 0.6245
#> iteration 69 : lambda 0.3753 0.6247
#> iteration 70 : lambda 0.3751 0.6249
#> iteration 71 : lambda 0.3749 0.6251
#> iteration 72 : lambda 0.3748 0.6252
#> iteration 73 : lambda 0.3746 0.6254
#> iteration 74 : lambda 0.3745 0.6255
#> iteration 75 : lambda 0.3743 0.6257
#> iteration 76 : lambda 0.3742 0.6258
#> iteration 77 : lambda 0.3741 0.6259
#> iteration 78 : lambda 0.374 0.626
#> iteration 79 : lambda 0.3738 0.6262
#> iteration 80 : lambda 0.3737 0.6263
#> iteration 81 : lambda 0.3736 0.6264
#> iteration 82 : lambda 0.3735 0.6265
#> iteration 83 : lambda 0.3735 0.6265
#> iteration 84 : lambda 0.3734 0.6266
#> iteration 85 : lambda 0.3733 0.6267
#> iteration 86 : lambda 0.3732 0.6268
#> iteration 87 : lambda 0.3731 0.6269
#> iteration 88 : lambda 0.3731 0.6269
#> iteration 89 : lambda 0.373 0.627
#> iteration 90 : lambda 0.373 0.627
#> iteration 91 : lambda 0.3729 0.6271
#> iteration 92 : lambda 0.3728 0.6272
#> iteration 93 : lambda 0.3728 0.6272
#> iteration 94 : lambda 0.3727 0.6273
#> iteration 95 : lambda 0.3727 0.6273
#> iteration 96 : lambda 0.3727 0.6273
#> iteration 97 : lambda 0.3726 0.6274
#> iteration 98 : lambda 0.3726 0.6274
#> iteration 99 : lambda 0.3725 0.6275
#> iteration 100 : lambda 0.3725 0.6275
#> iteration 101 : lambda 0.3725 0.6275
#> iteration 102 : lambda 0.3724 0.6276
#> iteration 103 : lambda 0.3724 0.6276
#> iteration 104 : lambda 0.3724 0.6276
#> iteration 105 : lambda 0.3724 0.6276
#> iteration 106 : lambda 0.3723 0.6277
#> iteration 107 : lambda 0.3723 0.6277
#> iteration 108 : lambda 0.3723 0.6277
#> iteration 109 : lambda 0.3723 0.6277
#> iteration 110 : lambda 0.3723 0.6277
#> iteration 111 : lambda 0.3722 0.6278
#> iteration 112 : lambda 0.3722 0.6278
#> iteration 113 : lambda 0.3722 0.6278
#> iteration 114 : lambda 0.3722 0.6278
#> iteration 115 : lambda 0.3722 0.6278
#> iteration 116 : lambda 0.3722 0.6278
#> iteration 117 : lambda 0.3722 0.6278
#> iteration 118 : lambda 0.3721 0.6279
#> iteration 119 : lambda 0.3721 0.6279
#> iteration 120 : lambda 0.3721 0.6279
#> iteration 121 : lambda 0.3721 0.6279
#> iteration 122 : lambda 0.3721 0.6279
#> iteration 123 : lambda 0.3721 0.6279
#> iteration 124 : lambda 0.3721 0.6279
#> iteration 125 : lambda 0.3721 0.6279
#> iteration 126 : lambda 0.3721 0.6279
#> iteration 127 : lambda 0.3721 0.6279
#> iteration 128 : lambda 0.372 0.628
#> iteration 129 : lambda 0.372 0.628
#> iteration 130 : lambda 0.372 0.628
#> iteration 131 : lambda 0.372 0.628
#> iteration 132 : lambda 0.372 0.628
#> iteration 133 : lambda 0.372 0.628
#> iteration 134 : lambda 0.372 0.628
#> iteration 135 : lambda 0.372 0.628
#> iteration 136 : lambda 0.372 0.628
#> iteration 137 : lambda 0.372 0.628
#> iteration 138 : lambda 0.372 0.628
#> iteration 139 : lambda 0.372 0.628
#> iteration 140 : lambda 0.372 0.628
#> iteration 141 : lambda 0.372 0.628
#> iteration 142 : lambda 0.372 0.628
#> iteration 143 : lambda 0.372 0.628
#> iteration 144 : lambda 0.372 0.628
#> iteration 145 : lambda 0.372 0.628
#> iteration 146 : lambda 0.372 0.628
#> iteration 147 : lambda 0.372 0.628
#> iteration 148 : lambda 0.372 0.628
#> iteration 149 : lambda 0.372 0.628
#> iteration 150 : lambda 0.372 0.628
#> iteration 151 : lambda 0.372 0.628
#> iteration 152 : lambda 0.372 0.628
#> iteration 153 : lambda 0.372 0.628
#> iteration 154 : lambda 0.372 0.628
#> iteration 155 : lambda 0.372 0.628
#> iteration 156 : lambda 0.372 0.628
#> iteration 157 : lambda 0.372 0.628
#> iteration 158 : lambda 0.372 0.628
#> iteration 159 : lambda 0.372 0.628
#> iteration 160 : lambda 0.3719 0.6281
#> iteration 161 : lambda 0.3719 0.6281
#> iteration 162 : lambda 0.3719 0.6281
#> iteration 163 : lambda 0.3719 0.6281
#> iteration 164 : lambda 0.3719 0.6281
#> iteration 165 : lambda 0.3719 0.6281
#> iteration 166 : lambda 0.3719 0.6281
#> iteration 167 : lambda 0.3719 0.6281
#> iteration 168 : lambda 0.3719 0.6281
#> iteration 169 : lambda 0.3719 0.6281
#> iteration 170 : lambda 0.3719 0.6281
#> iteration 171 : lambda 0.3719 0.6281
#> iteration 172 : lambda 0.3719 0.6281
#> iteration 173 : lambda 0.3719 0.6281
#> iteration 174 : lambda 0.3719 0.6281
#> iteration 175 : lambda 0.3719 0.6281
#> iteration 176 : lambda 0.3719 0.6281
#> iteration 177 : lambda 0.3719 0.6281
#> iteration 178 : lambda 0.3719 0.6281
#> iteration 179 : lambda 0.3719 0.6281
#> iteration 180 : lambda 0.3719 0.6281
#> iteration 181 : lambda 0.3719 0.6281
#> iteration 182 : lambda 0.3719 0.6281
#> iteration 183 : lambda 0.3719 0.6281
#> iteration 184 : lambda 0.3719 0.6281
#> iteration 185 : lambda 0.3719 0.6281
#> iteration 186 : lambda 0.3719 0.6281
#> iteration 187 : lambda 0.3719 0.6281
#> iteration 188 : lambda 0.3719 0.6281
#> iteration 189 : lambda 0.3719 0.6281
#> iteration 190 : lambda 0.3719 0.6281
#> iteration 191 : lambda 0.3719 0.6281
#> iteration 192 : lambda 0.3719 0.6281
#> iteration 193 : lambda 0.3719 0.6281
#> iteration 194 : lambda 0.3719 0.6281
#> iteration 195 : lambda 0.3719 0.6281
#> iteration 196 : lambda 0.3719 0.6281
#> iteration 197 : lambda 0.3719 0.6281
#> iteration 198 : lambda 0.3719 0.6281
#> iteration 199 : lambda 0.3719 0.6281
#> iteration 200 : lambda 0.3719 0.6281
#> iteration 201 : lambda 0.3719 0.6281
#> iteration 202 : lambda 0.3719 0.6281
#> iteration 203 : lambda 0.3719 0.6281
#> iteration 204 : lambda 0.3719 0.6281
#> iteration 205 : lambda 0.3719 0.6281
#> iteration 206 : lambda 0.3719 0.6281
#> iteration 207 : lambda 0.3719 0.6281
#> iteration 208 : lambda 0.3719 0.6281
#> iteration 209 : lambda 0.3719 0.6281
#> iteration 210 : lambda 0.3719 0.6281
#> iteration 211 : lambda 0.3719 0.6281
#> iteration 212 : lambda 0.3719 0.6281
#> iteration 213 : lambda 0.3719 0.6281
#> iteration 214 : lambda 0.3719 0.6281
#> iteration 215 : lambda 0.3719 0.6281
#> iteration 216 : lambda 0.3719 0.6281
#> iteration 217 : lambda 0.3719 0.6281
#> iteration 218 : lambda 0.3719 0.6281
#> iteration 219 : lambda 0.3719 0.6281
#> iteration 220 : lambda 0.3719 0.6281
#> iteration 221 : lambda 0.3719 0.6281
#> iteration 222 : lambda 0.3719 0.6281
#> iteration 223 : lambda 0.3719 0.6281
#> iteration 224 : lambda 0.3719 0.6281
#> iteration 225 : lambda 0.3719 0.6281
#> iteration 226 : lambda 0.3719 0.6281
#> iteration 227 : lambda 0.3719 0.6281
#> iteration 228 : lambda 0.3719 0.6281
#> iteration 229 : lambda 0.3719 0.6281
#> iteration 230 : lambda 0.3719 0.6281
#> iteration 231 : lambda 0.3719 0.6281
#> iteration 232 : lambda 0.3719 0.6281
#> iteration 233 : lambda 0.3719 0.6281
#> iteration 234 : lambda 0.3719 0.6281
#> iteration 235 : lambda 0.3719 0.6281
#> iteration 236 : lambda 0.3719 0.6281
#> iteration 237 : lambda 0.3719 0.6281
#> iteration 238 : lambda 0.3719 0.6281
#> iteration 239 : lambda 0.3719 0.6281
#> iteration 240 : lambda 0.3719 0.6281
#> iteration 241 : lambda 0.3719 0.6281
#> iteration 242 : lambda 0.3719 0.6281
#> iteration 243 : lambda 0.3719 0.6281
#> iteration 244 : lambda 0.3719 0.6281
#> # iter 244, lambda 0.3719 0.6281, total time 148.43 s
g5 <- ggplot() +
geom_point(aes(sce$detected, sce$subsets_mito_percent, colour = mix2d$posterior[, 2])) +
scale_colour_viridis() +
ggtitle("2d gaussian mixture")
g6 <- ggplot() +
geom_point(aes(sce$detected, sce$subsets_mito_percent, colour = mix2d$posterior[, 2] > 0.8)) +
ggtitle("2d gaussian mixture")
cowplot::plot_grid(g5, g6)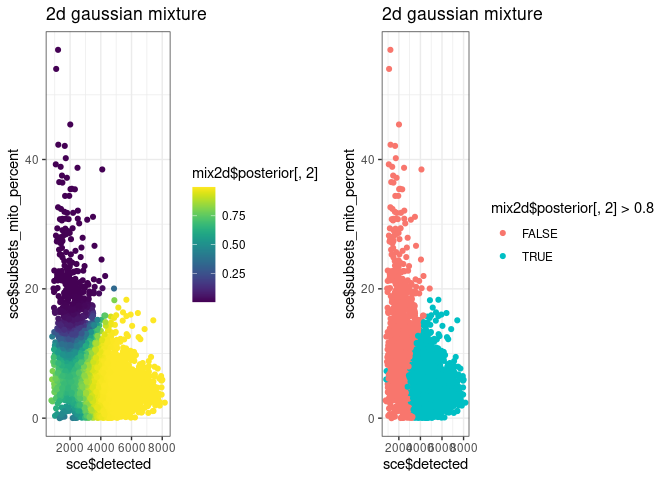
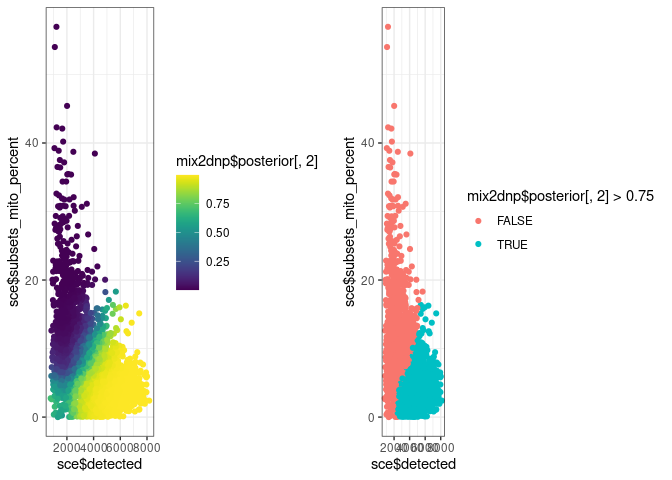
g7 <- ggplot() +
geom_point(aes(sce$detected, sce$subsets_mito_percent, colour = mix2dnp$posterior[, 2])) +
scale_colour_viridis()
g8 <- ggplot() +
geom_point(aes(sce$detected, sce$subsets_mito_percent, colour = mix2dnp$posterior[, 2] > 0.75))
cowplot::plot_grid(g7, g8)