# from py to R ------------------------------------------------------------
# to convert the a python notebook to Rmd file
# with https://pkgs.rstudio.com/rmarkdown/reference/convert_ipynb.html
# for one sample ----------------------------------------------------------
set.seed(123)
rnorm(n = 25,mean = 100,sd = 20)
#> [1] 88.79049 95.39645 131.17417 101.41017 102.58575 134.30130 109.21832
#> [8] 74.69878 86.26294 91.08676 124.48164 107.19628 108.01543 102.21365
#> [15] 88.88318 135.73826 109.95701 60.66766 114.02712 90.54417 78.64353
#> [22] 95.64050 79.47991 85.42218 87.49921
# (1) generate multiple random samples ----------------------------------------
library(tidyverse)
# with https://gist.github.com/avallecam/ee009a8065921e74054935f87e3bf7d5
data <- tibble(
class = 1:10,
n = 25, # this changes
mean = 100,
sd = 20
) %>%
mutate(sample = pmap(select(., -class), rnorm))
data
#> # A tibble: 10 × 5
#> class n mean sd sample
#> <int> <dbl> <dbl> <dbl> <list>
#> 1 1 25 100 20 <dbl [25]>
#> 2 2 25 100 20 <dbl [25]>
#> 3 3 25 100 20 <dbl [25]>
#> 4 4 25 100 20 <dbl [25]>
#> 5 5 25 100 20 <dbl [25]>
#> 6 6 25 100 20 <dbl [25]>
#> 7 7 25 100 20 <dbl [25]>
#> 8 8 25 100 20 <dbl [25]>
#> 9 9 25 100 20 <dbl [25]>
#> 10 10 25 100 20 <dbl [25]>
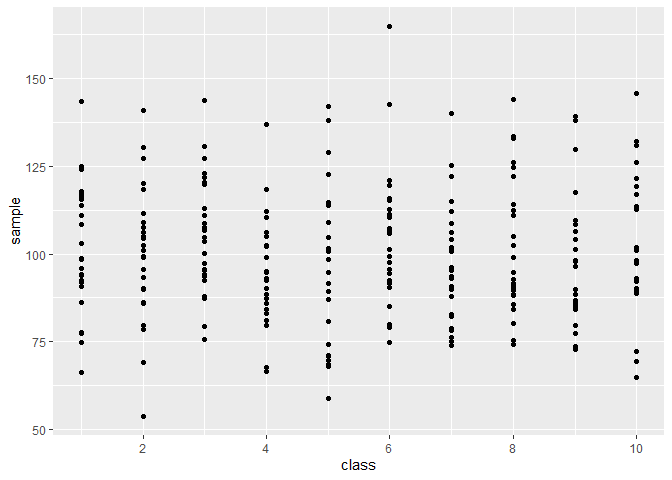
## distribution of values per sample ---------------------------------------
# with https://stackoverflow.com/questions/15622001/how-to-display-only-integer-values-on-an-axis-using-ggplot2
data %>%
select(class,sample) %>%
unnest(sample) %>%
ggplot(aes(x = class, y = sample)) +
geom_point() +
scale_x_continuous(breaks= scales::pretty_breaks())## (2) create summary statistics per sample ------------------------------------
# with https://dplyr.tidyverse.org/articles/rowwise.html#motivation
data_summary <-
data %>%
select(class, sample) %>%
rowwise() %>%
mutate(s_mean = mean(sample),
s_sd = sd(sample)) %>%
ungroup()
data_summary
#> # A tibble: 10 × 4
#> class sample s_mean s_sd
#> <int> <list> <dbl> <dbl>
#> 1 1 <dbl [25]> 102. 18.4
#> 2 2 <dbl [25]> 100. 19.4
#> 3 3 <dbl [25]> 106. 16.6
#> 4 4 <dbl [25]> 94.5 15.6
#> 5 5 <dbl [25]> 95.4 23.6
#> 6 6 <dbl [25]> 104. 19.8
#> 7 7 <dbl [25]> 97.2 17.0
#> 8 8 <dbl [25]> 102. 19.5
#> 9 9 <dbl [25]> 97.5 18.5
#> 10 10 <dbl [25]> 103. 20.1
## distribution of all summary statistics ----------------------------------
data_summary %>%
ggplot(aes(x = s_mean)) +
geom_histogram()
#> `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.## (3) variability of sample means ---------------------------------------------
data_summary %>%
summarise(
n_samples = n(),
mean_s_mean = mean(s_mean),
sd_s_mean = sd(s_mean))
#> # A tibble: 1 × 3
#> n_samples mean_s_mean sd_s_mean
#> <int> <dbl> <dbl>
#> 1 10 100. 3.89
# multiple samples for multiple sizes -------------------------------------
# with https://tidyr.tidyverse.org/reference/expand.html
expand_summary <-
# (0) generate multiple combinations, instead of tibble
expand_grid(
n = c(10,25,50), # this changes
class = 1:1000,
mean = 100,
sd = 20
) %>%
# (1) generate multiple random samples
mutate(sample = pmap(select(., -class), rnorm)) %>%
# (2) create summary statistics per sample
select(n, class, sample) %>%
rowwise() %>%
mutate(s_mean = mean(sample),
s_sd = sd(sample)) %>%
ungroup()
# distribution of all summary statistics
expand_summary %>%
ggplot(aes(x = s_mean)) +
geom_histogram() +
facet_grid(~n)
#> `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.expand_summary %>%
# (3) variability of sample means
group_by(n) %>%
summarise(
n_samples = n(),
mean_s_mean = mean(s_mean),
sd_s_mean = sd(s_mean))
#> # A tibble: 3 × 4
#> n n_samples mean_s_mean sd_s_mean
#> <dbl> <int> <dbl> <dbl>
#> 1 10 1000 100. 6.26
#> 2 25 1000 99.9 3.87
#> 3 50 1000 100. 2.77
# mean of sample means
# standard deviation of sample means ~ standard errorCreated on 2023-11-13 with reprex v2.0.2



figure 1