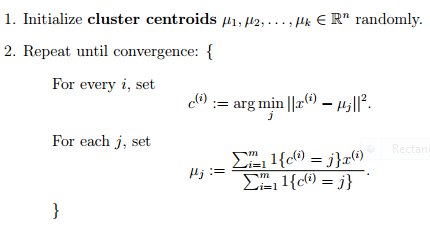
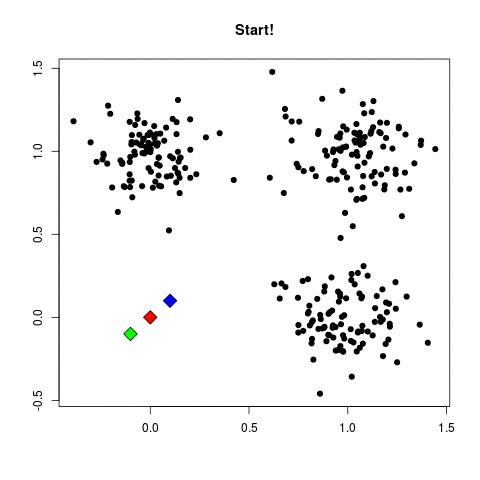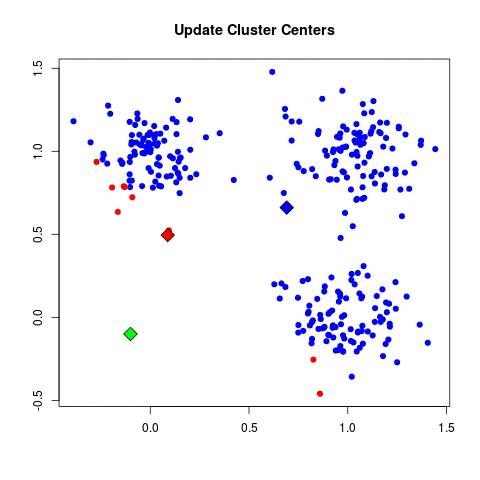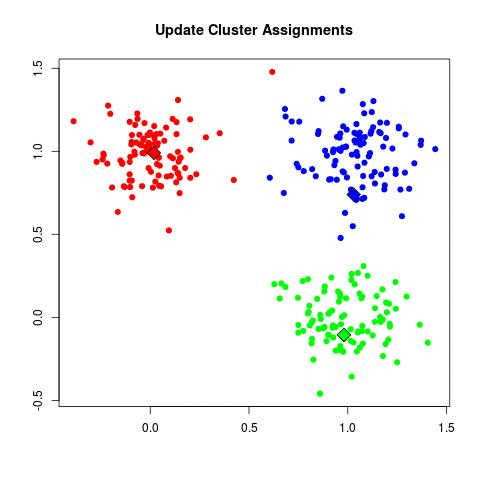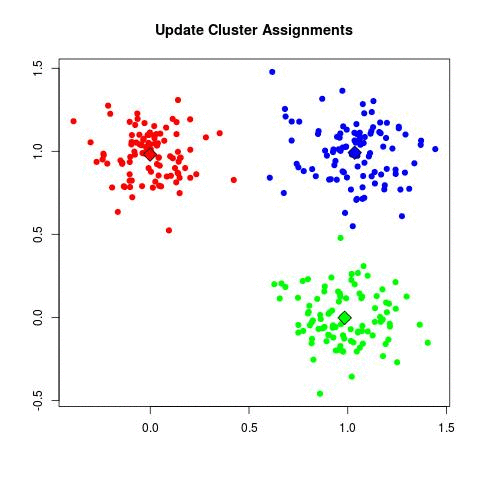
- Unsupervised
- Determines the center of clusters
- Given how many clusters you want to find
- Your data is numeric. It doesn't work with categorical features. We're computing the distance between real numbers!
- If you don't have labels for your data
- K-means is the simplest. To implement and to run. All you need to do is choose "k" and run it a number of times.
- K-means and other clustering algorithms shine when you have multivariate data. They will "work" with 1-dimensional data, but they are not very smart anymore.
- useful when you have an idea of how many clusters actually exists in your space.
- Security
- Classification
- Generally identifying something unknown
- Place all the points
- Place centroids in random location
- for each point assign to the nearest centroid
Let P be any point in the plane of a triangle with vertices A, B, and C and centroid G. Then the sum of the squared distances of P from the three vertices exceeds the sum of the squared distances of the centroid G from the vertices by three times the squared distance between P and G:
The sum of the squares of the triangle's sides equals three times the sum of the squared distances of the centroid from the vertices:
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.animation as animation
def load_dataset(name):
return np.loadtxt(name)
def euclidian(a, b):
return np.linalg.norm(a-b)
def plot(dataset, history_centroids, belongs_to):
colors = ['r', 'g']
fig, ax = plt.subplots()
for index in range(dataset.shape[0]):
instances_close = [i for i in range(len(belongs_to)) if belongs_to[i] == index]
for instance_index in instances_close:
ax.plot(dataset[instance_index][0], dataset[instance_index][1], (colors[index] + 'o'))
history_points = []
for index, centroids in enumerate(history_centroids):
for inner, item in enumerate(centroids):
if index == 0:
history_points.append(ax.plot(item[0], item[1], 'bo')[0])
else:
history_points[inner].set_data(item[0], item[1])
print("centroids {} {}".format(index, item))
plt.pause(0.8)
def kmeans(k, epsilon=0, distance='euclidian'):
history_centroids = []
if distance == 'euclidian':
dist_method = euclidian
dataset = load_dataset('durudataset.txt')
# dataset = dataset[:, 0:dataset.shape[1] - 1]
num_instances, num_features = dataset.shape
prototypes = dataset[np.random.randint(0, num_instances - 1, size=k)]
history_centroids.append(prototypes)
prototypes_old = np.zeros(prototypes.shape)
belongs_to = np.zeros((num_instances, 1))
norm = dist_method(prototypes, prototypes_old)
iteration = 0
while norm > epsilon:
iteration += 1
norm = dist_method(prototypes, prototypes_old)
prototypes_old = prototypes
for index_instance, instance in enumerate(dataset):
dist_vec = np.zeros((k, 1))
for index_prototype, prototype in enumerate(prototypes):
dist_vec[index_prototype] = dist_method(prototype,
instance)
belongs_to[index_instance, 0] = np.argmin(dist_vec)
tmp_prototypes = np.zeros((k, num_features))
for index in range(len(prototypes)):
instances_close = [i for i in range(len(belongs_to)) if belongs_to[i] == index]
prototype = np.mean(dataset[instances_close], axis=0)
# prototype = dataset[np.random.randint(0, num_instances, size=1)[0]]
tmp_prototypes[index, :] = prototype
prototypes = tmp_prototypes
history_centroids.append(tmp_prototypes)
# plot(dataset, history_centroids, belongs_to)
return prototypes, history_centroids, belongs_to
def execute():
dataset = load_dataset('durudataset.txt')
centroids, history_centroids, belongs_to = kmeans(2)
plot(dataset, history_centroids, belongs_to)
execute()0.196705753183788 0.266174967499455
0.413286989521383 0.355828352990633
0.338435546719209 0.435738258997923
0.103801517189990 0.164344805836115
0.159052363075132 0.325059012698889
0.0669054926780630 0.487418074001379
0.335731444739015 0.0379836806470678
0.285495537731203 0.293509583541386
0.0848835330132443 0.206943248886680
0.0738278885758684 0.154568213233134
0.238039859133728 0.131917020763398
0.454051208253475 0.379383132540102
0.276087513357917 0.497607990564876
0.0164699463749383 0.0932857220706846
0.0269314632177781 0.390572634267382
0.402531614279451 0.0978989905133660
0.225687427351724 0.496179486589963
0.191323114779979 0.401130784882144
0.394821851844845 0.212113354951653
0.182143434749897 0.364431934025687
1.49835358252355 1.40350138880436
1.80899026719904 1.93497908617805
1.35650893348105 1.47948454563248
1.07324343448981 1.23179161166312
1.59099145527485 1.39629024850978
1.91018783072814 1.70507747511279
1.19376593616661 1.55855903456055
1.43236779153440 1.75663070089437
1.74915972906801 1.99548105855526
1.03918448664758 1.96243140436663
1.94632498980548 1.53506710525616
1.76367332366376 1.96387012997171
1.55882055050956 1.11562587918126
1.18384294446577 1.05144829323021
1.49794881501895 1.30434894563657
1.51784560023405 1.58019183314271
1.99424301064405 1.53096445233828
1.85485168309068 1.90120809265314
1.96240393971197 1.54055042517024
1.67894100897703 1.43198061085668