Created
May 18, 2018 21:11
-
-
Save grovduck/29cae3c00749eaa715a9ba7e9b0cfd64 to your computer and use it in GitHub Desktop.
This file contains hidden or bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| class FeatureSpace(object): | |
| def __init__(self, max_k): | |
| self.max_k = max_k | |
| self.knn_classifier = KNeighborsClassifier(max_k) | |
| def train(self): | |
| raise NotImplementedError | |
| def transform(self, raw_scores): | |
| raise NotImplementedError | |
| def k_neighbors(self, scores): | |
| return self.knn_classifier.kneighbors(scores) | |
| class CCAFeatureSpace(FeatureSpace): | |
| def __init__(self, max_k, n_axes, spp_transform=None): | |
| super(CCAFeatureSpace, self).__init__(n_axes, max_k) | |
| self.n_axes = n_axes | |
| self.spp_transform = spp_transform | |
| def train(self, species_df, environmental_df, id_field='ID'): | |
| # Here's where you run NumpyCCA | |
| # Set a bunch of self variables that define the space | |
| self.fcids = np.array(species_df.__dict__[id_field]) | |
| spp_arr = np.array(species_df.drop(columns=[id_field])) | |
| env_arr = np.array(enironmental_df.drop(columns=[id_field])) | |
| cca_obj = NumpyCCA(spp_arr, env_arr) | |
| self.coefficients = cca_obj.coefficients() | |
| self.lc_scores = cca_obj.site_lc_scores() | |
| # ... | |
| # At the end, set the plot_scores on the knn_classifier | |
| self.knn_classifier.fit(plot_scores, fcids) | |
| def transform(self, raw_scores): | |
| # This is function transform_env_score in current code | |
| return transform_env_score(raw_scores) | |
| def k_neighbors(self, scores): | |
| dist, ind = super(CCAFeatureSpace, self).k_neighbors(scores) | |
| neighbor_ids = self.fcids[ind[0]] | |
| return dist[0], neighbor_ids | |
| class EuclideanFeatureSpace(FeatureSpace): | |
| def __init__(self, max_k): | |
| super(EuclideanFeatureSpace, self).__init__(max_k) | |
| def train(self, environmental_df, id_field='ID') | |
| self.fcids = np.array(environmental_df.__dict__[id_field]) | |
| # Here's where you'd set up the axes based on normalized | |
| # environmental variables, like in our first example | |
| # ... | |
| # At the end, set the plot_scores on the knn_classifier | |
| self.knn_classifier.fit(plot_scores, fcids) | |
| def transform(self, raw_scores): | |
| # Standardize environmental variables | |
| # Not currently implemented | |
| return subtract_mean_divide_std(raw_scores) | |
| def k_neighbors(self, scores): | |
| # TODO: Redundant with CCA because self.fcids is member | |
| # of subclasses | |
| dist, ind = super(EuclideanFeatureSpace, self).k_neighbors(scores) | |
| neighbor_ids = self.fcids[ind[0]] | |
| return dist[0], neighbor_ids | |
| # A class to represent a stack (bands) of rasters that is capable | |
| # of imputation and attribution | |
| class RasterList(object): | |
| def __init__(self, rasters): | |
| # Build the raster list | |
| self.raster_list = # ... | |
| def extract(self, point_feature_class): | |
| # Extract spatial signatures at plot locations | |
| return pd.DataFrame(env_signatures) | |
| def impute(self, feature_space_obj): | |
| # This is essentially your updatePixels loop from | |
| # lines 347-351 | |
| pix_array_dim = pix_array.shape | |
| num_squares_x = pix_array_dim[1] | |
| num_squares_y = pix_array_dim[2] | |
| nn_neighbors = np.ones((band_count, num_squares_x, num_squares_y)) | |
| for num_x in range(0, int(num_squares_x)): | |
| for num_y in range(0, int(num_squares_y)): | |
| raw_score = pix_array[:, num_x, num_y]) | |
| transformed_score = feature_space_obj.transform(raw_score) | |
| self.distances, self.neighbors = ( | |
| feature_space_obj.k_neighbors(transformed_score) | |
| ) | |
| def map_attribute(self, attr_df, map_attr, k=None, distance_weighting=None): | |
| # Yet to do, but basically the logic in lines 351:353 | |
| def main(): | |
| # Set up your input layers | |
| # This is more like pseudocode, but I'm guesing you have methods | |
| # to do this stuff | |
| raster_stack = RasterList([ | |
| Raster('C:/path/to/raster/file/1'), | |
| Raster('C:/path/to/raster/file/2'), | |
| Raster.extract_band('C:/path/to/harmonic/regression', 1) | |
| Raster.extract_band('C:/path/to/harmonic/regression', 2) | |
| # ... | |
| ]) | |
| # Sample point feature class | |
| fc = FeatureClass('C:/path/to/feature/class') | |
| # Get the environmental matrix | |
| env_df = raster_stack.extract(fc) | |
| # We'll pretend we're running a CCA, so read in a species file | |
| # as well | |
| spp_df = pd.read_csv('C:/path/to/species/file') | |
| # Create the CCA object and train it using the two matrices | |
| fs_obj = CCAFeatureSpace(8, 10, spp_transform='SQRT') | |
| fs_obj.train(env_df, spp_df, id_field='FCID') | |
| # Get all neighbors and distances using the requested feature space | |
| # Note that we're guaranteed that the same ordering of environmental | |
| # data that went into the feature space creation is the same when | |
| # running impute | |
| raster_stack.impute(fs_obj) | |
| # We'll pretend we have a separate CSV file of attributes, although | |
| # they could be connected to the point feature class as well | |
| attr_df = pd.read_csv('C:/path/to/attributes/file') | |
| # Map a given variable and return a raster | |
| psme_raster = raster_stack.map_attribute( | |
| attr_df, 'PSME', k=5, distance_weighting='INVERSE' | |
| ) |
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment
@grovduck I am refactoring pieces of the CCA-kNN-Attribute mapping so that each gets it's own function. One they have their own functions, they can be chained together. Take a look at the example screenshot:
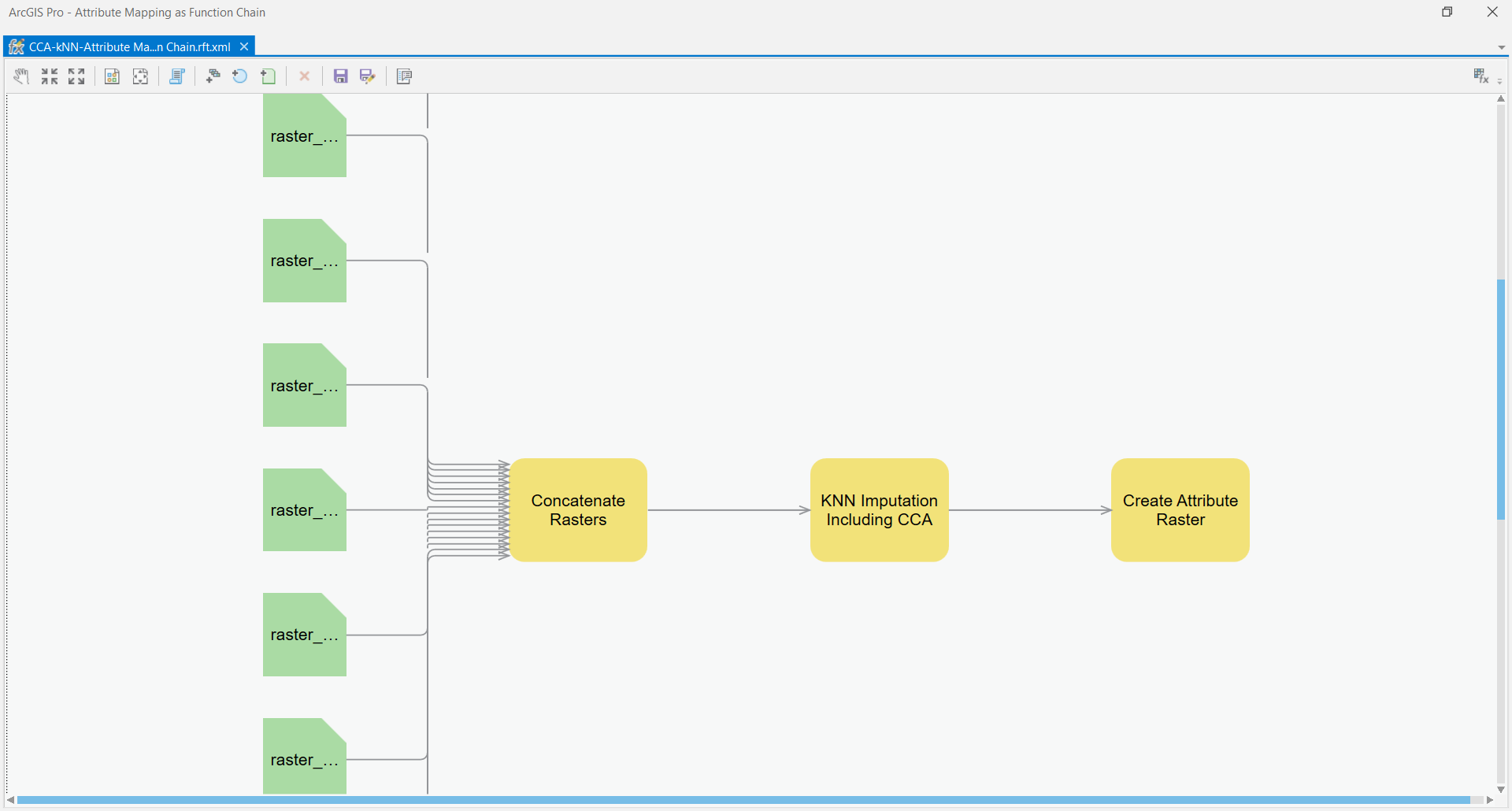
This is how Esri intends for multiple processes to be chained together. What do you think of this model? Any luck exploring the Python Raster Functions on your machine?