|
==================================== |
|
https://github.com/ray-project/ray/issues/31384 |
|
Title: [<Ray component: Core>] @ray.remote function runs on head node instead of worker node |
|
Body ### What happened + What you expected to happen |
|
|
|
I run a python function on ray cluster with @ray.remote, but the function runs on head node instead of worker node
|
|
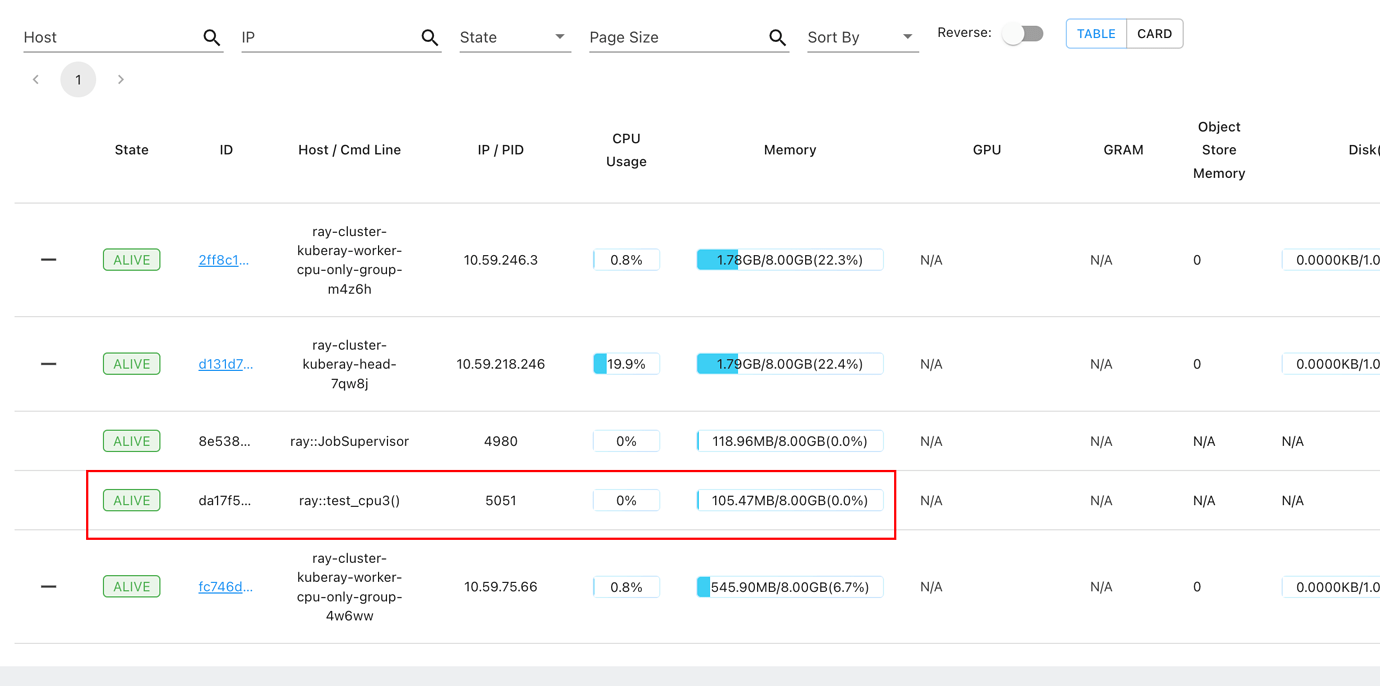
|
|
|
|
|
|
### Versions / Dependencies |
|
|
|
Ray version: 2.0.0 |
|
|
|
### Reproduction script |
|
|
|
```
|
|
import ray
|
|
|
|
@ray.remote
|
|
def test_cpu3():
|
|
import time
|
|
time.sleep(150)
|
|
return "done"
|
|
|
|
my_task = test_cpu3.remote()
|
|
ray.get(my_task)
|
|
``` |
|
|
|
### Issue Severity |
|
|
|
None |
|
Comments: Functions are scheduled on a node that has available cpus. So it is normal it is scheduled on a worker node. If you'd like to avoid it setting `--num-cpus` of a head node as 0 (`ray start --head --num-cpus=0`) or use the node affinity scheduling to avoid scheduling on a head node https://docs.ray.io/en/master/ray-core/scheduling/index.html#nodeaffinityschedulingstrategy |
|
thanks a lot, it works |
|
------------------------------------ |
|
When will you run into this issue: |
|
This issue will occur when running a python function on ray cluster with @ray.remote and the function runs on head node instead of worker node. |
|
|
|
Here's how to fix it: |
|
Functions are scheduled on a node that has available CPUs. So it is normal that it is scheduled on a worker node. However, if you'd like to avoid scheduling the function on a head node, you can set the `--num-cpus` of a head node as 0 when starting Ray: `ray start --head --num-cpus=0`. Alternatively, you can use the node affinity scheduling strategy to avoid scheduling on a head node. |
|
|
|
Explanation: |
|
By default, Ray distributes workloads on nodes with available CPUs. The head node typically has available CPU resources and may run the function by default. To avoid running the function on the head node, you can set the `--num-cpus` of the head node as 0 when starting Ray. This will prevent workloads from being scheduled on the head node. Alternatively, you can use the node affinity scheduling strategy to specify which nodes should be used for particular workloads so that you can ensure that workloads are not scheduled on head nodes. |
|
==================================== |
|
==================================== |
|
https://github.com/ray-project/ray/issues/26173 |
|
Title: [clusters][docker] Docker `ports` configuration in ray cluster configuration file. |
|
Body ### Description |
|
|
|
In configuration documentation https://docs.ray.io/en/latest/cluster/config.html#docker
|
|
there seems to be no `port` type.
|
|
|
|
I would love to know how to connect to ray dashboard for ray clusters running in docker. Many thanks! |
|
|
|
### Use case |
|
|
|
_No response_ |
|
Comments: The docker config is hardcoded to share a network with the host.
|
|
|
|
To temporarily port-forward or expose to a single machine, you can use the `ray dashboard` command to forward the connection.
|
|
|
|
You'll need to expose your dashboard port/add port forwarding to expose it publicly (this is cloud-provider specific, but you need to allow inbound connections on port 8265 in your aws security group for example) |
|
@spacegoing - Can you add more specific info to this issue to help our engineers? Command you used? Error message? |
|
You may also want to add `--dashboard-host=0.0.0.0` to the `ray start` command to allow connections from outside the host. |
|
@spacegoing - Can you try Alex's and Ian's comments? Did this solve it for you? Please let us know! Thanks! |
|
@christy Hi Christy, sorry for the late reply. Everything works now. Many thanks for everybody's help!
|
|
|
|
Here is the ray cluster configuration file [gpu.yaml](https://github.com/spacegoing/ray_test/blob/master/gpu.yaml) I configured ray cluster to use p2.xlarge (it has 1 GPU / instance).
|
|
|
|
I run following commands:
|
|
1. On my `local` laptop, connecting to an AWS server through ssh port forwarding:
|
|
`ssh -L "8265:localhost:8265" user@myserver`
|
|
2. On `myserver`, spin up the ray cluster by:
|
|
`ray up gpu.yaml`
|
|
3. On `myserver`, connecting to the ray cluster's dashboard by:
|
|
`ray dashboard gpu.yaml`
|
|
The output is:
|
|
```
|
|
Attempting to establish dashboard locally at localhost:8265 connected to remote port 8265
|
|
2022-07-11 22:33:06,977 INFO util.py:335 -- setting max workers for head node type to 0
|
|
2022-07-11 22:33:07,128 VWARN commands.py:319 -- Loaded cached provider configuration from /tmp/ray-config-5252fb2b960c8410b8834d876bb9584c1850dd74
|
|
2022-07-11 22:33:07,128 WARN commands.py:327 -- If you experience issues with the cloud provider, try re-running the command with --no-config-cache.
|
|
2022-07-11 22:33:07,129 VINFO utils.py:146 -- Creating AWS resource `ec2` in `us-west-2`
|
|
2022-07-11 22:33:07,248 VINFO utils.py:146 -- Creating AWS resource `ec2` in `us-west-2`
|
|
2022-07-11 22:33:07,363 INFO command_runner.py:394 -- Fetched IP: 54.244.188.177
|
|
2022-07-11 22:33:07,364 INFO log_timer.py:30 -- NodeUpdater: i-08886b554018827bc: Got IP [LogTimer=0ms]
|
|
2022-07-11 22:33:07,364 INFO command_runner.py:524 -- Forwarding ports
|
|
2022-07-11 22:33:07,364 VINFO command_runner.py:531 -- Forwarding port 8265 to port 8265 on localhost.
|
|
2022-07-11 22:33:07,364 VINFO command_runner.py:552 -- Running `None`
|
|
2022-07-11 22:33:07,364 VVINFO command_runner.py:555 -- Full command is `ssh -tt -L 8265:localhost:8265 -i ./aws_ray_pt_head.pem -o StrictHostKeyChecking=no -o UserKnownHostsFile=/dev/null -o IdentitiesOnly=yes -o ExitOnForwardFailure=yes -o ServerAliveInterval=5 -o ServerAliveCountMax=3 -o ControlMaster=auto -o ControlPath=/tmp/ray_ssh_1d41c853af/4315291b87/%C -o ControlPersist=10s -o ConnectTimeout=120s ubuntu@54.244.188.177 while true; do sleep 86400; done`
|
|
Warning: Permanently added '54.244.188.177' (ECDSA) to the list of known hosts.
|
|
```
|
|
5. On `local` laptop, visiting `localhost:8265`.
|
|
Here is a screenshot:
|
|
|
|
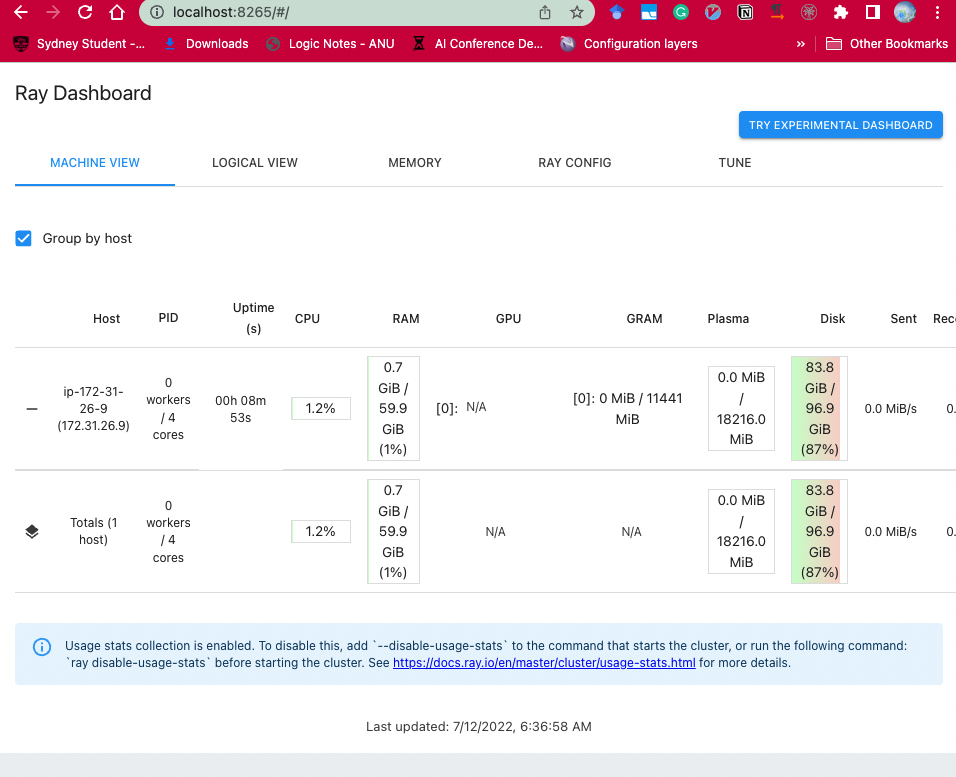
|
|
|
|
|
|
Just double-checking:
|
|
Did you have to connect to a remote server to `ray up` as a work-around to the issue, or were you working with a remote server when you originally posted this issue? |
|
@DmitriGekhtman Hi Dmitri, the first time I post I used the server. My laptop is an MBP with apple M1 chip. |
|
------------------------------------ |
|
When will you run into this issue? |
|
|
|
You will run into this issue when you are trying to connect to the Ray dashboard for Ray clusters running in Docker. |
|
|
|
Here's how to fix it. |
|
|
|
To temporarily port-forward or expose to a single machine, use the `ray dashboard` command to forward the connection. You need to expose your dashboard port/add port forwarding to expose it publicly. For instance, if you want to allow inbound connections on port 8265 in your AWS security group, do so. |
|
|
|
You may also want to add `--dashboard-host=0.0.0.0` to the `ray start` command to allow connections from outside the host. |
|
|
|
Here is an example of port-forwarding to connect to the Ray dashboard: |
|
|
|
1. On your local laptop, connect to an AWS server through ssh port forwarding: |
|
`ssh -L "8265:localhost:8265" user@myserver` |
|
|
|
2. On the server, spin up the Ray cluster by running: |
|
`ray up gpu.yaml` |
|
|
|
3. On the server, connect to the Ray cluster's dashboard by running: |
|
`ray dashboard gpu.yaml` |
|
|
|
4. On your local laptop, visit `localhost:8265`. |
|
|
|
Explanation: |
|
|
|
The docker configuration is hardcoded to share a network with the host. You need to port-forward or expose your dashboard port to connect to the Ray dashboard for Ray clusters running in Docker. |
|
|
|
By running the `ray dashboard` command and port-forwarding with `ssh`, the connection will be forwarded to `localhost:8265`. You can then visit `localhost:8265` on your local laptop to see the Ray dashboard. |
|
|
|
Furthermore, you can also add `--dashboard-host=0.0.0.0` to the `ray start` command to allow connections from outside the host. |
|
==================================== |
|
==================================== |
|
https://github.com/ray-project/ray/issues/33179 |
|
Title: [data] Dataset.num_blocks() not always == len(Dataset.input_files()) |
|
Body ### Description
|
|
|
|
Hi! I'd like to read a parquet dataset from a directory of files on local disk where each file corresponds exactly to one block in the dataset. The docs suggest that this should be the case —
|
|
|
|
> Read Parquet files into a tabular Dataset. The Parquet data will be read into [Arrow Table](https://arrow.apache.org/docs/python/generated/pyarrow.Table.html) blocks. Although this simple example demonstrates reading a single file, note that Datasets can also read directories of Parquet files, with one tabular block created per file.
|
|
|
|
— but I keep running into situations where `Dataset.num_blocks() != len(Dataset.input_files())`. I can modify the number of blocks by tweaking `DatasetContext`'s `block_splitting_enabled` and `target_max_block_size` values. I delved into the source code but couldn't find where the disconnect between input files and output blocks was occurring. (I got as far as the `BlockOutputBuffer` before tapping out...)
|
|
|
|
I'm not doing anything unusual with the `ray.data.read_parquet(dir_path)` call, so I'm assuming this is an expected feature, and the problem is actually misleading info in the linked documentation. Please let me know if there's a way to guarantee 1:1 file:block reading of parquet data! If not, clarifying in the docs when that relationship doesn't hold would be a help.
|
|
|
|
### Link
|
|
|
|
https://docs.ray.io/en/latest/data/creating-datasets.html#supported-file-formats |
|
Comments: Hey @bdewilde, thanks for opening an issue!
|
|
|
|
Looks like this an inaccuracy in our docs. I've opened a PR to fix: https://github.com/ray-project/ray/pull/33185.
|
|
|
|
To achieve 1:1 file-to-block reading, you could try setting `parallelism` to the number of files:
|
|
|
|
```
|
|
ray.data.read_parquet(..., parallelism=NUM_FILES)
|
|
```
|
|
|
|
Also, just of out curiosity, why're you interested in a 1:1 mapping between files blocks? |
|
Hi @bveeramani , thanks for confirming! I'll try setting `parallelism` as you suggest, I didn't realize that would guarantee the 1:1 mapping between file and batch.
|
|
|
|
Each file represents an independent chunk of data from a much larger dataset that I've already grouped using AWS Athena. I'd like to process each chunk via `Dataset.map_batches(batch_size=None)` or, possibly, `BatchMapper(batch_size=None).transform(ds)`. I could in principle re-group the full dataset via `Dataset.groupby()`, but it's a bit clunky — it's grouped on multiple columns, which ray doesn't support, so requires workarounds — and slow, since the full dataset is very large. It's much faster to read in N files at a time, process each independently, then write the results back to disk in a more streaming fashion. |
|
------------------------------------ |
|
When will you run into this issue? |
|
|
|
You may run into this issue if you are trying to read a parquet dataset from a directory of files on local disk where each file corresponds exactly to one block in the dataset. You may notice that `Dataset.num_blocks() != len(Dataset.input_files())`. |
|
|
|
Here's how to fix it: |
|
|
|
To achieve 1:1 file-to-block reading, you can set `parallelism` to the number of files. For example, if you have three files, you can set `parallelism` to three as shown below: |
|
|
|
```python |
|
ray.data.read_parquet(dir_path, parallelism=3) |
|
``` |
|
|
|
Explanation: |
|
|
|
As per the conversation in the issue, the problem is actually misleading information in the documentation. To achieve the desired behavior, you can set the `parallelism` argument in the `read_parquet` method to the number of files you have. This guarantees 1:1 mapping between file and batch, which should solve the issue. |
|
|
|
Each file represents an independent chunk of data from a much larger dataset that may have already been grouped. It is much faster to read in N files at a time, process each independently, then write the results back to disk in a more streaming fashion than to group the full dataset via `Dataset.groupby()`. |
|
==================================== |