Last active
September 13, 2023 13:30
-
-
Save vankesteren/0467c4ccdb333ce73cbefc75a58d9c58 to your computer and use it in GitHub Desktop.
Conformal prediction for linear regression and random forest. NB: pretty ugly and slow code.
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| # conformal prediction intervals for linear regression and random forest | |
| library(tidyverse) | |
| library(pbapply) | |
| library(parallel) | |
| # fully conformal prediction | |
| conformal_quantile <- function(x, y, x_new, y_new, frm) { | |
| N <- length(x) | |
| df <- tibble(x = c(x, x_new), y = c(y, y_new)) | |
| fy <- lm(frm, df) | |
| scores <- residuals(fy)^2 | |
| # find the quantile of y_new score | |
| return(ecdf(scores[1:N])(scores[N+1])) | |
| } | |
| conformal_interval <- function(x, y, x_new, frm, level = 0.95, resolution = 0.1, min_y = mean(x) - IQR(x)*3, max_y = mean(x) + IQR(x)*3) { | |
| y_new <- seq(min_y, max_y, resolution) | |
| y_qtl <- vapply(y_new, conformal_quantile, x = x, y = y, x_new = x_new, frm = frm, FUN.VALUE = 0.) | |
| out <- if (all(y_qtl >= level)) c(-Inf, Inf) else range(y_new[y_qtl < level]) | |
| names(out) <- c("lwr", "upr") | |
| return(out) | |
| } | |
| # tryout | |
| set.seed(456) | |
| N <- 1000 | |
| x <- rnorm(N) + 2 | |
| f <- function(x) 1+x-2*x^2+0.3*x^3 | |
| y <- f(x) + rnorm(N) | |
| plot(x, y) | |
| df <- tibble(x = x, y = y) | |
| frm <- y ~ poly(x, 3) | |
| fit <- lm(frm, df) | |
| x_new <- seq(min(x) - 0.1, max(x) + 0.1, 0.1) | |
| pred <- predict(fit, data.frame(x = x_new), interval = "pred", level = 0.95) | |
| conf <- t(pbsapply(x_new, conformal_interval, x = x, y = y, min_y = -23, max_y = 9, res = 0.1, frm = frm)) | |
| colnames(conf) <- c("lwr_c", "upr_c") | |
| as_tibble(pred) |> | |
| bind_cols(conf) |> | |
| mutate(x = x_new) |> | |
| ggplot(aes(x = x, y = fit)) + | |
| geom_ribbon(aes(ymin = lwr, ymax = upr), fill = "#ababab") + | |
| geom_ribbon(aes(ymin = lwr_c, ymax = upr_c), fill = "#ab12ab") + | |
| geom_line() + | |
| geom_point(data = tibble(x = x, fit = y)) | |
| # tryout with ranger | |
| conformal_quantile_rf <- function(x, y, x_new, y_new, frm) { | |
| N <- length(x) | |
| df <- tibble(x = c(x, x_new), y = c(y, y_new)) | |
| fy <- ranger(frm, df) | |
| scores <- (df$y - predict(fy, df)$predictions)^2 | |
| # find the quantile of y_new score | |
| return(ecdf(scores[1:N])(scores[N+1])) | |
| } | |
| conformal_interval_rf <- function(x, y, x_new, frm, level = 0.95, resolution = 0.1, min_y = mean(x) - IQR(x)*3, max_y = mean(x) + IQR(x)*3) { | |
| y_new <- seq(min_y, max_y, resolution) | |
| y_qtl <- vapply(y_new, conformal_quantile_rf, x = x, y = y, x_new = x_new, frm = frm, FUN.VALUE = 0.) | |
| out <- if (all(y_qtl >= level)) c(-Inf, Inf) else range(y_new[y_qtl < level]) | |
| names(out) <- c("lwr", "upr") | |
| return(out) | |
| } | |
| frm <- y ~ x | |
| fit <- ranger(frm, df) | |
| x_new <- seq(min(x) - 0.1, max(x) + 0.1, 0.1) | |
| pred <- predict(fit, data.frame(x = x_new))$prediction | |
| cl <- makeCluster(10) | |
| clusterExport(cl, c("conformal_interval_rf", "conformal_quantile_rf", "x", "y", "frm")) | |
| clusterEvalQ(cl, { | |
| library(ranger) | |
| library(tibble) | |
| }) | |
| conf <- pbsapply(x_new, | |
| function(xn) conformal_interval_rf( | |
| x = x, | |
| y = y, | |
| x_new = xn, | |
| frm = frm, | |
| resolution = 0.1, | |
| min_y = -23, | |
| max_y = 9 | |
| ), | |
| cl = cl | |
| ) | |
| stopCluster(cl) | |
| rownames(conf) <- c("lwr_c", "upr_c") | |
| tibble(fit = pred) |> | |
| bind_cols(t(conf)) |> | |
| mutate(x = x_new, lwr_c = smooth(lwr_c), upr_c = smooth(upr_c)) |> | |
| ggplot(aes(x = x, y = fit)) + | |
| geom_ribbon(aes(ymin = lwr_c, ymax = upr_c), fill = "#ab12ab") + | |
| geom_line() + | |
| geom_point(data = tibble(x = x, fit = y)) |
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment
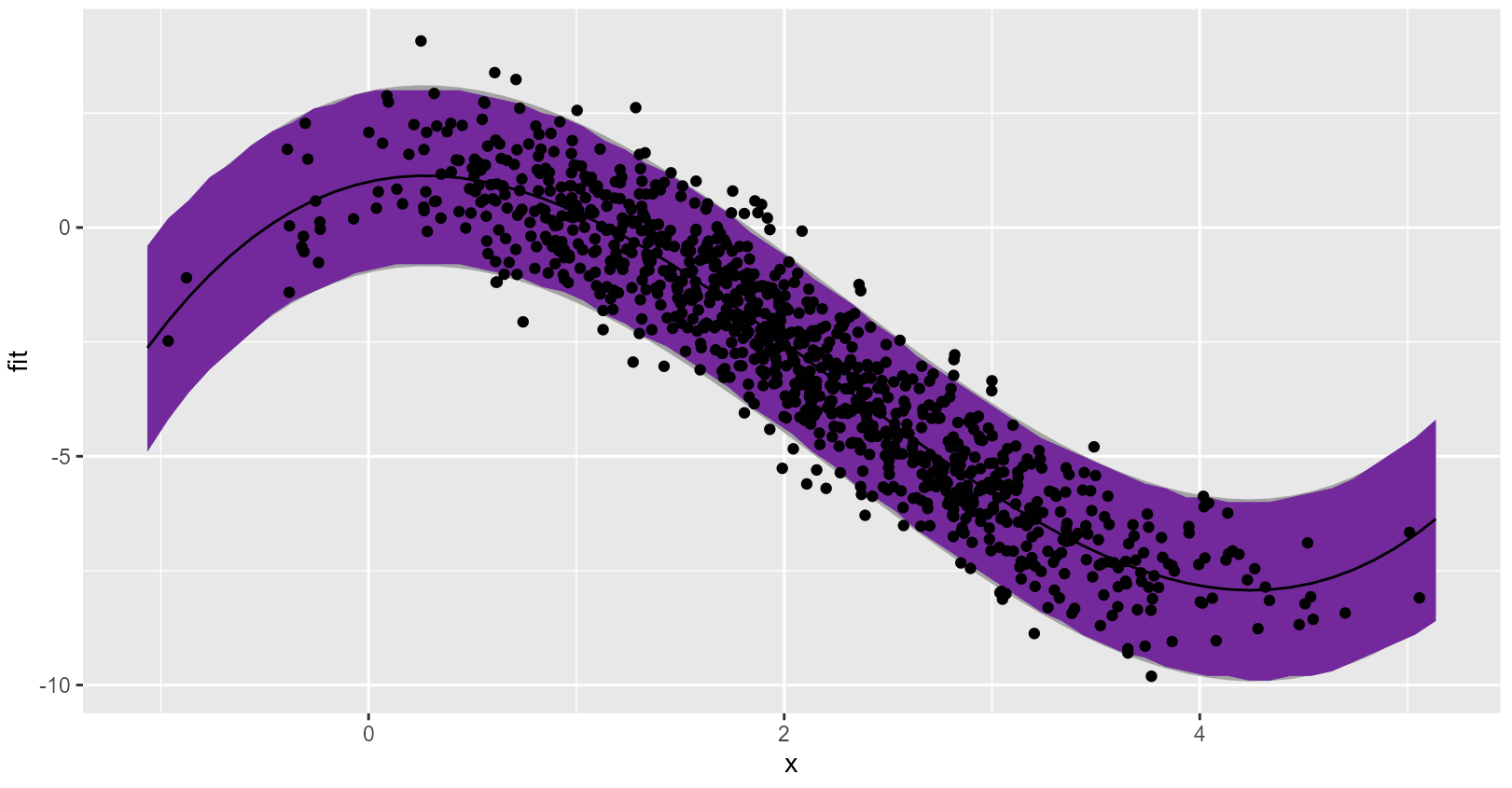
Linear regression prediction interval looks a lot like the standard interval, which is good because data were generated and modeled with all the correct assumptions:
Random forest smoothed prediction interval is computationally very intensive, but possible! See here: