-
-
Save MLWave/4a3f8b0fee43d45646cf118bda4d202a to your computer and use it in GitHub Desktop.
| # Author: HJ van Veen <[email protected]> | |
| # Description: Experiment to learn a tSNE transformer for new | |
| # test data with a multi-output GBM | |
| # | |
| # Idea first seen at lvdmaaten.github.io/tsne | |
| # > [...] it is not possible to embed test points in an existing | |
| # > map [...] | |
| # > A potential approach to deal with this would be to train | |
| # > a multivariate regressor to predict the map location from | |
| # > the input data. | |
| # | |
| # Part of code adapted from Fabian Pedregosa, Olivier Grisel, | |
| # Mathieu Blondel, Gael Varoquaux, | |
| # originally licensed under "BSD 3 clause (C) INRIA 2011". | |
| from sklearn import (manifold, datasets, preprocessing, model_selection, | |
| decomposition, metrics, multioutput) | |
| from xgboost import XGBRegressor | |
| import matplotlib.pyplot as plt | |
| import numpy as np | |
| # For data we use 6 different digit classes of 8x8 pixels | |
| digits = datasets.load_digits(n_class=6) | |
| X = digits.data # (1083, 64) | |
| y = digits.target # (1083, ) | |
| # Split the data into 66% train and 33% test set. | |
| X_train, X_test, y_train, y_test = model_selection.train_test_split(X, | |
| y, | |
| test_size=0.33, | |
| random_state=0) | |
| # First, PCA 2-D (which has .transform()) to illustrate and evaluate | |
| lens = decomposition.PCA(n_components=2, random_state=0) | |
| X_lens_train = lens.fit_transform(X_train) | |
| X_lens_test = lens.transform(X_test) | |
| # Normalize the lens within 0-1 | |
| scaler = preprocessing.MinMaxScaler() | |
| X_lens_train = scaler.fit_transform(X_lens_train) | |
| X_lens_test = scaler.transform(X_lens_test) | |
| # Fit a model and predict the lens values from the original features | |
| model = XGBRegressor(n_estimators=2000, max_depth=20, learning_rate=0.01) | |
| model = multioutput.MultiOutputRegressor(model) | |
| model.fit(X_train, X_lens_train) | |
| preds = model.predict(X_test) | |
| # Evaluate exhaustively | |
| print("PREDICTION\t\tGROUND TRUTH") | |
| for p, g in zip(preds, X_lens_test): | |
| print(p, g) | |
| print("MAE", metrics.mean_absolute_error(X_lens_test, preds)) | |
| # Now TSNE (which has no .transform()) and a visual evaluation | |
| lens = manifold.TSNE(n_components=2, init='pca', random_state=0) | |
| X_lens_train = lens.fit_transform(X_train) | |
| # Normalize the lens within 0-1 | |
| X_lens_train = scaler.fit_transform(X_lens_train) | |
| # Fit a model and predict the lens values from the original features | |
| model.fit(X_train, X_lens_train) | |
| X_tsne = model.predict(X_test) | |
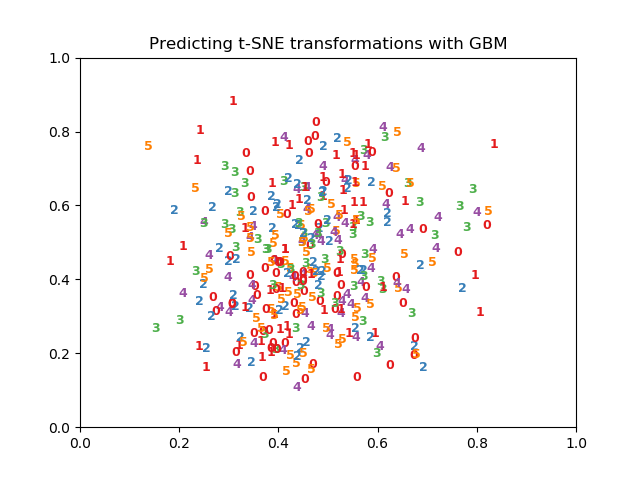
| # Visualize predictions | |
| plt.figure() | |
| for i in range(X_tsne.shape[0]): | |
| plt.text(X_tsne[i, 0], X_tsne[i, 1], str(y_test[i]), | |
| color=plt.cm.Set1(y_test[i] / 10.), | |
| fontdict={'weight': 'bold', 'size': 9}) | |
| plt.title("Predicting t-SNE transformations with GBM") | |
| plt.savefig("tsne-predictions.png") |
I'm not sure if using an XGBRegressor here is appropriate as you may run into extrapolation using your new data, and tree based methods in general do not perfom well with extrapolation. This post explains the problem: http://ellisp.github.io/blog/2016/12/10/extrapolation
I suspect you are right. XGBoost scores very well on accuracy, not so much on extrapolation. In further local experiments I switched to a MLP and see, like the great post you linked, the extrapolation being better. Also switched to Standard Scaler as this does not give a hard cap between 0-1 for values lower or higher than seen in train set, like MinMaxScaler does.
If you want to do this right, then create a neural net and optimize the t-SNE loss directly, like in: Learning a Parametric Embedding by Preserving Local Structure [pdf]
In my computer (PC running Ubuntu with python 3.0) tsne-transform.py is performing much worse than
as shown in the figure at https://twitter.com/MLWave/status/882153224492527616 The following is the figure I get when running tsne-transform.py
I also tried the modified code provided by @arm_gilles to visualize errors and I can neither obtain a well separated signal as shown in https://twitter.com/arm_gilles/status/882220762328858624 The following is what I get when I run the modified tsne-transform.py by @arm_gilles
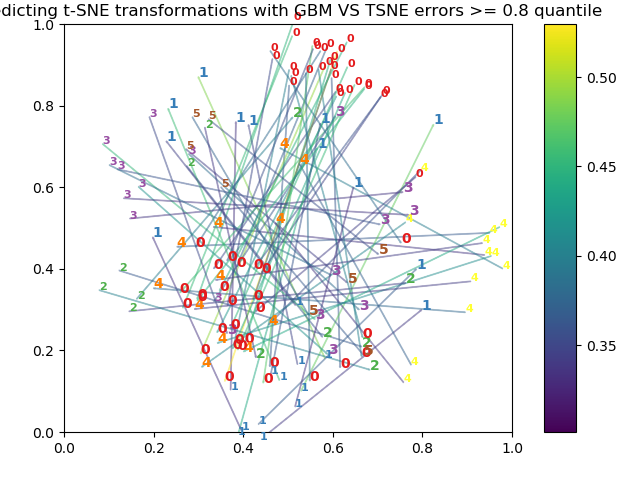
Any idea of what could be the problem?
I tried both pieces of codes many times and always get poor results. I also tried them on different Linux computers without luck.
Thanks, Joaquin
If you have this kind of error :
Then you can fix it with :
Source Scikit Github issue