To select N leaves.
- Pick a leaf at random. Add it to a selected list.
While N < len(selected):
- For all not selected leaves, compute minimum distance (d) to any leaf in selected.
- Identiy the leaf that has maximum d, and add this to selected.
Last active
February 4, 2019 10:41
-
-
Save davipatti/b450918a8eb800f17628969327040623 to your computer and use it in GitHub Desktop.
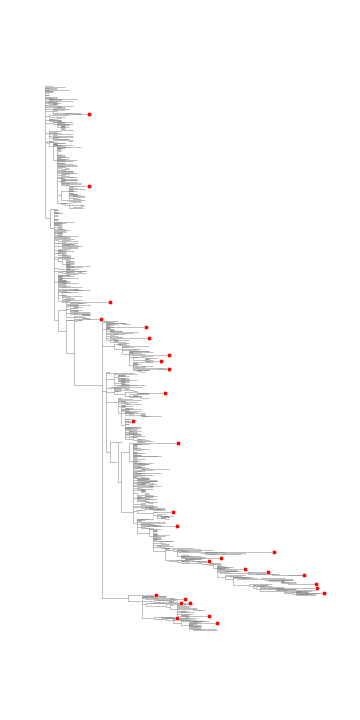
Select phylogenetically distant taxa in a tree
This file contains hidden or bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| def selectMostDistantLeaves(tree, n): | |
| """Select n phylogenetically distance leaves in a tree. | |
| Args: | |
| tree (dendropy.Tree) | |
| n (int) | |
| Returns: | |
| list containing labels of leaf nodes | |
| """ | |
| tree = Tree(tree) # distancesFromNode alters tree, so make a copy | |
| node = random.choice(tree.leaf_nodes()) | |
| selected = pd.DataFrame({node: distancesFromNode(tree, node)}) | |
| while selected.shape[1] < n: | |
| node = selected.apply(min, axis=1).idxmax() # Could store only minima, not entire df. Would save repeated computation | |
| selected[node] = distancesFromNode(tree, node) | |
| return [n.taxon.label for n in selected.columns] | |
| def distancesFromNode(tree, node): | |
| """Compute distances from all nodes in tree to node. | |
| Args: | |
| tree (dendropy.Tree) | |
| node (dendropy.node) Must be node in tree. | |
| Returns: | |
| pd.Series containing distance from node | |
| """ | |
| tree.reroot_at_node(node) | |
| series = pd.Series(tree.calc_node_root_distances(), index=tree.leaf_nodes()) | |
| series[node] = 0 | |
| return series |
Author
davipatti
commented
Feb 4, 2019
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment