-
-
Save mirko77/3f4a101cd4a77e2ae3e760d44d18d901 to your computer and use it in GitHub Desktop.
| library(httr) | |
| library(jsonlite) # if needing json format | |
| cID<-"999" # client ID | |
| secret<- "F00HaHa00G" # client secret | |
| proj.slug<- "YourProjectSlug" # project slug | |
| form.ref<- "YourFormRef" # form reference | |
| branch.ref<- "YourFromRef+BranchExtension" # branch reference | |
| res <- POST("https://five.epicollect.net/api/oauth/token", | |
| body = list(grant_type = "client_credentials", | |
| client_id = cID, | |
| client_secret = secret)) | |
| http_status(res) | |
| token <- content(res)$access_token | |
| # url.form<- paste("https://five.epicollect.net/api/export/entries/", proj.slug, "?map_index=0&form_ref=", form.ref, "&format=json", sep= "") ## if using json | |
| url.form<- paste("https://five.epicollect.net/api/export/entries/", proj.slug, "?map_index=0&form_ref=", form.ref, "&format=csv&headers=true", sep= "") | |
| res1<- GET(url.form, add_headers("Authorization" = paste("Bearer", token))) | |
| http_status(res1) | |
| # ct1<- fromJSON(rawToChar(content(res1))) ## if using json | |
| ct1<- read.csv(res1$url) | |
| str(ct1) | |
| # url.branch<- paste("https://five.epicollect.net/api/export/entries/", proj.slug, "?map_index=0&branch_ref=", branch.ref, "&format=json&per_page=1000", sep= "") ## if using json; pushing max number of records from default 50 to 1000 | |
| url.branch<- paste("https://five.epicollect.net/api/export/entries/", proj.slug, "?map_index=0&branch_ref=", branch.ref, "&format=csv&headers=true", sep= "") | |
| res2<- GET(url.branch, add_headers("Authorization" = paste("Bearer", token))) | |
| http_status(res2) | |
| ct2<- read.csv(res2$url) | |
| # ct2<- fromJSON(rawToChar(content(res2))) ## if using json | |
| str(ct2) |
Hi @mirko77 - I do have filenames that are the correct format that I have queried using the code you originally posted above. Sorry I was not clear in my comment that I was simplifying for the sake of the example! I also studied the JavaScript prior to my question two days ago, and just now I tried to fork and test the fiddle you linked above based on your suggestion. I saved what I tested in the fork (I think) and left a few comments as to my thought process, but could not get the JS to work; unfortunately, my JavaScript skills are not that great (I use R for most things).
I will try to give a full picture of what I am doing in R:
- First, in EpiCollect via a browser, I created an 'app' and obtained a 'Client ID' and 'Client Secret' as you demonstrate helpfully above.
- Second, I define my private project slug and export entries from my private project into R via the API using the token method you demonstrate above. This works no problem. I get a token that I name
tokenthat is a very long string of random letters, numbers, and symbols. - Third, to use httr::GET() to obtain a photo, I use this code:
projectSlug <- "meier-cider-trees" # Actual name of my private project
treePhoto <- "7ba0da82-7088-4fe1-88cf-2b86d8d9e8b0_1576430696.jpg" # Actual file name from row 1 of my private project
writeFile <- "export_treePhoto.jpg" # Example name of file written to local destination
imageURL <- paste0("https://five.epicollect.net/api/export/media/", projectSlug, "?type=photo?format=entry_original&name=", treePhoto)
httr::GET(imageURL, add_headers("Authorization" = paste("Bearer", token)), write_disk(path = writeFile, overwrite = TRUE))
I then get the following server response:
Response [https://five.epicollect.net/api/export/media/meier-cider-trees?type=photo?format=entry_original&name=7ba0da82-7088-4fe1-88cf-2b86d8d9e8b0_1576430696.jpg]
Date: 2020-03-22 17:26
Status: 400
Content-Type: application/vnd.api+json; charset=utf-8
Size: 144 B
<ON DISK> export_treePhoto.jpg
As you can see, I think my file names are the correct format, and the Status: 400 message indicates a bad request that is either not understandable or is missing required parameters. So far, I have not been able to figure out either a) what is not understandable in the URL I am sending, or b) what parameters are missing from the httr::GET() request.
I hope that I have been clearer, and thank you for your time so far!
I am not an R expert, but if you can get the image for a public project (just set up a test one) having an error on the private one is just an authentication issue
@schafnoir I'm doing the exact same thing as you (as in retrieving media from a private's project), and followed the same steps.
Using the exacts same code as you did, I obtained the same error on export.
But I found a typo in your code above form imageURL. You need to replace ?type=photo?format by ?type=photo&format and it works!
Hope it helps!
Best,
Steph
@StephPeriquet - I believe you are correct about the misplaced ? in my code, thank you for the eagle eyes! Unfortunately, I am unable to test since I am now receiving a '404' error when I try to query for data entries using code that worked a couple weeks ago, and I need the data entries to get the image file names. Not sure what's going on, but one step forward and a few back, it seems.
Cheers,
Courtney
Hi Courtney,
Sorry to hear that... I download the csv on my machine then use it to extract image names, might be a solution?
Best,
Steph
@mirko77 - I wanted to let you know that in the code chunk above, the following no longer works due to the new per_page=1000 limit imposed on the API:
url.branch<- paste("https://five.epicollect.net/api/export/entries/", proj.slug, "?map_index=0&branch_ref=", branch.ref, "&format=json&per_page=1000000", sep= "")
Cheers,
Courtney
@mirko77 - I wanted to let you know that in the code chunk above, the following no longer works due to the new
per_page=1000limit imposed on the API:
url.branch<- paste("https://five.epicollect.net/api/export/entries/", proj.slug, "?map_index=0&branch_ref=", branch.ref, "&format=json&per_page=1000000", sep= "")Cheers,
Courtney
per_page=1000000 would crash even a Google server, just get the data in multiple requests using the page parameter.
You can also reduce the number of entries on each request by using the filter_from and filter_to parameters, to get just the new entries.
@mirko77 - no argument from me as to the wisdom of paginated requests! The code I quoted immediately above came from your example at the top of the page, and all I was pointing out is that it doesn't work, so you might want to update the example :-)
@mirko77 - no argument from me as to the wisdom of paginated requests! The code I quoted immediately above came from your example at the top of the page, and all I was pointing out is that it doesn't work, so you might want to update the example :-)
Ah ok, understood.
The code has not been written by myself ;)
@schafnoir thanks for reporting it, now it should be fixed!
Hello,
this line actually gives a error
ct1<- read.csv(res1$url)
Error in file(file, "rt") : cannot open the connection
In addition: Warning message:
In file(file, "rt") :
cannot open URL 'https://five.epicollect.net/api/export/entries/cccu-needle-collection?map_index=0&form_ref=3fc893ee3bd341ad831e8239be29d633_5cabc23cb7fa9&format=csv&per_page=1000&page=1': HTTP status was '404 Not Found'
what should I do?
Hello there,
I am sorry I have no idea how to sort out this issue...
You would need to share your whole code so we can have a look. It looks like the project you are trying to reach doesn't exists...
Best,
Steph
if you go to https://five.epicollect.net/api/export/entries/cccu-needle-collection?map_index=0&form_ref=3fc893ee3bd341ad831e8239be29d633_5cabc23cb7fa9&format=csv&per_page=1000&page=1 with your browser you will get an access denied error since that project is private.
You need authorization to get entries for a public project, please see https://developers.epicollect.net/api-authentication/create-client-app
Hello there,
I am sorry I have no idea how to sort out this issue...
You would need to share your whole code so we can have a look. It looks like the project you are trying to reach doesn't exists...Best,
Steph
That is interesting. I used exactly the code from above. The Jason one works fine. But the CSV one works fine until the ct1<- read.csv(res1$url).
Hello there,
I am sorry I have no idea how to sort out this issue...
You would need to share your whole code so we can have a look. It looks like the project you are trying to reach doesn't exists...Best,
Steph
if you go to https://five.epicollect.net/api/export/entries/cccu-needle-collection?map_index=0&form_ref=3fc893ee3bd341ad831e8239be29d633_5cabc23cb7fa9&format=csv&per_page=1000&page=1 with your browser you will get an
access denied errorsince that project is private.
You need authorization to get entries for a public project, please see https://developers.epicollect.net/api-authentication/create-client-app
That is exactly what I did. I tried using Python, which works fine. I used exactly the code from above, the Jason one works fine but the CSV one showed error until the ct1<- read.csv(res1$url).
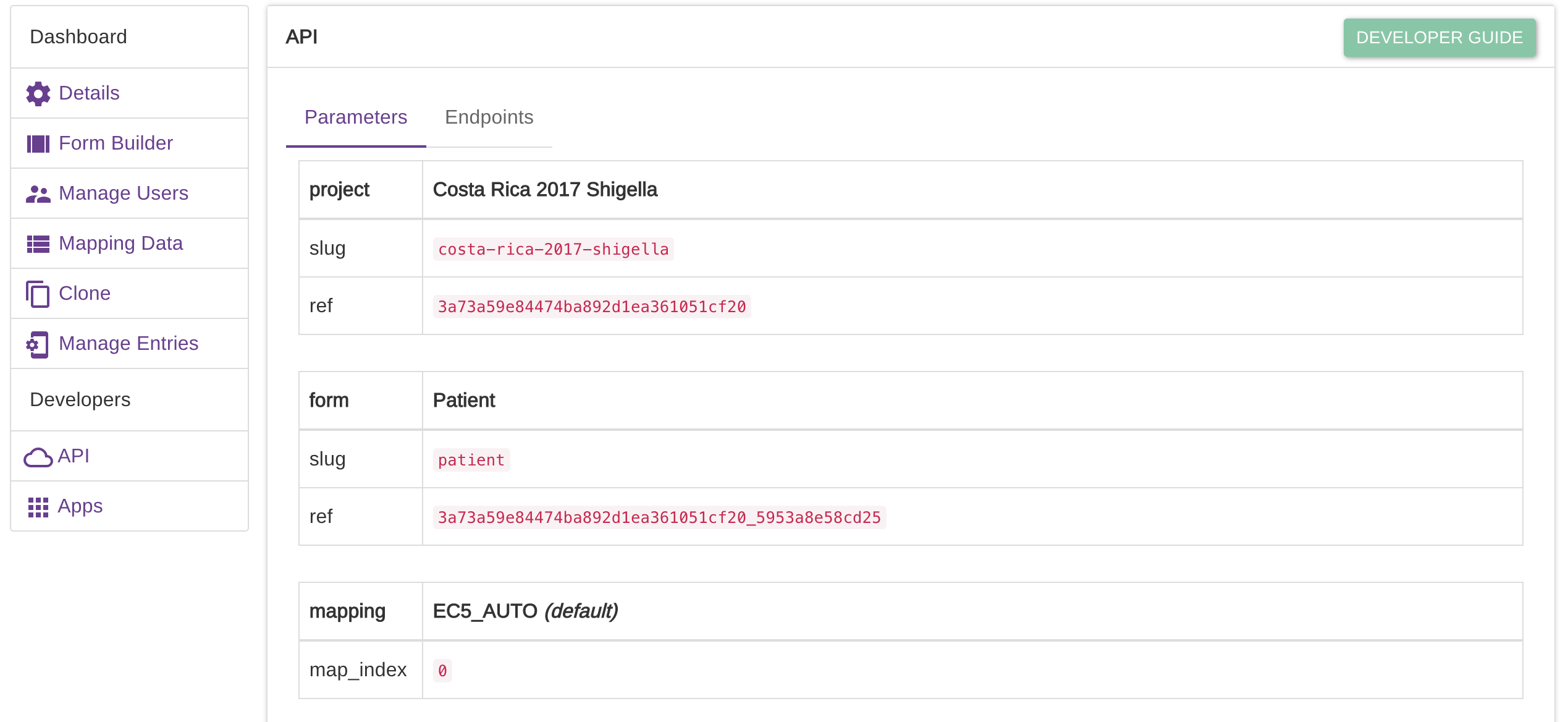
Hi @mirko77 - I was just wondering where I could find the 'project.slug' and 'form.ref' information from Epicollect? I'm guessing those respond to the name at the end of the project URL (https://five.epicollect.net/project/project.slug) and the form name in the Form Builder, respectively? I'm able to retrieve the token successfully when I run the code above, but get errors with the part of the code beginning with 'url.form:'
http_status(res1)
$category
[1] "Client error"
$reason
[1] "Bad Request"
$message
[1] "Client error: (400) Bad Request"
Any thoughts on what could be causing this?
Thanks,
Jay
@jaymwin - if you click on the 'Details' button associated with any of your EpiCollect projects, you should see an 'API' link on the left of the page that then opens. Click 'API' and you will then see the parameters you are after.
@schafnoir answer is correct.
If you have only one form, the form ref parameter is not even needed, as it will default to the first form anyway.
thanks @schafnoir and @mirko77 - using those parameters and removing form.ref here did the trick:
url.form<- paste("https://five.epicollect.net/api/export/entries/", proj.slug, "?map_index=0&form_ref=", "&format=csv&headers=true", sep= "")
hey,
I am trying to set up an automated transfer of pics to my drive using AWS. Some days back it was working fine and it seems like it broke in the meantime. The problem is that it is not downloading original files but just the file that is named the same but has only 77B of information for each file. Any ideas? I have used the same code as presented above.

The code:
for( pic in pics){
imageURL <- paste0("https://five.epicollect.net/api/export/media/", proj.slug, "?type=photo&format=entry_original&name=", pic)
httr::GET(
imageURL,
add_headers("Authorization" = paste("Bearer", token)),
write_disk(
path = here("dta/img/", pic),
overwrite = TRUE)
)
## Upload on Google drive
# googledrive::drive_upload(media = here("dta/img/", pic),
# path = folder_id$id
# )
## Remove from local folder
# file.remove(here("dta/img/", pic))
Sys.sleep(2)
}
Example image URL is:
"https://five.epicollect.net/api/export/media/soyscout?type=photo&format=entry_original&name=7149429a-3eb3-4999-b106-d836090e6b59_1652127690.jpg"
We have no experience of AWS we afraid.
It looks like your requests get capped at 78b, so they are either wrong or you are over a quota (30 requests per minute for media files on our API =>https://developers.epicollect.net/#rate-limiting). You might want to catch any network errors as well. The 78b might be the error response, which you are saving as an image regardless of its content.
Is the access token valid? They expire every 2 hours. https://developers.epicollect.net/api-authentication/retrieve-token
You could try the Javascript example here, probably easier to debug things -> https://developers.epicollect.net/examples/getting-media
There is a fiddle to play with as well https://jsfiddle.net/mirko77/y45brprq/
Apologies for the delay- it was an error message. I would imagine it was happening because of too many calls when I was developing this as it disappeared once I left it all for a day or two and added time breaks between API calls. Thank you!
Great, I am glad the issue is solved!
That is interesting. I used exactly the code from above. The Jason one works fine. But the CSV one works fine until the ct1<- read.csv(res1$url).
@jimer666 I am facing the same issue you had, I am able to get the entries in the JSON format but the csv does not work. Were you able to resolve this issue?
Thank you
This line gives the error
ct1<- read.csv(res1$url)
Error in file(file, "rt") : cannot open the connection

photoName.jpgis not a valid file name for Epicollect5, they are saved in the following format02a48b70-6c46-11ea-bd3f-6fc459407ff6_1584885865.jpgLook at your entries export to get the right filenames
Try to fork this fiddle and use your credentials. Do not forget to uncomment the auth part https://jsfiddle.net/mirko77/y45brprq/