Last active
November 1, 2021 20:11
-
-
Save rafapereirabr/0374a22fea871b8d2268642f949d9c60 to your computer and use it in GitHub Desktop.
Calculate the ratiation model in R
This file contains hidden or bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| #' Code by Rafael H. M. Pereira and Marcus Saraiva | |
| #' | |
| #' Radiation model (Simini et al 2012) | |
| #' Tij = Ti * ((Pi * Pj) / ((Pi + Pij) * (Pi + Pj + Pij))) | |
| #' | |
| #' Where: | |
| #' | |
| #' Tij: Avarage flux between origin i and destination j predicted by the radiation model | |
| #' Ti: Proportion of population in i relative to total population | |
| #' Pi: Absolute population in i | |
| #' Pj: Absolute population in j | |
| #' Pij: Absolute population potentially reacheable from i under the travel | |
| #' necessary to tavel between i and j, excluding the population in i and j | |
| #' | |
| #' Reference: | |
| #' Simini, F., González, M. C., Maritan, A., & Barabási, A. L. (2012). A universal | |
| #' model for mobility and migration patterns. Nature, 484(7392), 96-100. | |
| #' | |
| ### load libraries -------------------------------------------------------- | |
| library(r5r) | |
| library(ggplot2) | |
| library(dplyr) | |
| library(data.table) | |
| library(pbapply) | |
| library(furrr) | |
| library(future) | |
| library(ggpointdensity) | |
| library(viridisLite) | |
| options(scipen = 999) | |
| ### calculate travel tim matrix -------------------------------------------------------- | |
| # build transport network | |
| data_path <- system.file("extdata/poa", package = "r5r") | |
| r5r_core <- setup_r5(data_path = data_path, temp_dir = TRUE) | |
| # load origin/destination points | |
| points <- read.csv(file.path(data_path, "poa_hexgrid.csv")) | |
| # keep only points with population | |
| points <- subset(points, population > 0) | |
| # calculate travel time matrix | |
| tt <- travel_time_matrix(r5r_core, | |
| origins = points, | |
| destinations = points, | |
| mode = "WALK", | |
| max_trip_duration = 200) | |
| head(points) | |
| head(tt) | |
| ### calculate components of the radiation model -------------------------------------------------------- | |
| tt[points, on=c('fromId'='id'), pop_orig := i.population ] | |
| tt[points, on=c('toId'='id'), pop_dest := i.population ] | |
| head(tt) | |
| tail(tt) | |
| # total population | |
| pop_total <- sum(points$population) | |
| # Share of total population in each origin | |
| tt[, Ti := pop_orig / pop_total, by = fromId] | |
| # total pop_orig accessible from origin give max the travel time between o-d pair | |
| # excluding the pop in O and D | |
| # part 1 | |
| # get unique combinations of origin and travel time | |
| a <- tt[, .(fromId, travel_time )] | |
| a <- unique(a) | |
| a[, row := .I] | |
| part1_fun <- function(i, ...){ # i = 50000 | |
| orig <- a$fromId[i] | |
| tlimit <- a$travel_time[i] | |
| temp <- setDT(tt)[ fromId==orig, .(pij_part1 = sum(pop_dest[which(travel_time <= tlimit)])), by= fromId ] | |
| temp$travel_time <- tlimit | |
| return(temp) | |
| } | |
| future::plan(strategy = 'multisession') | |
| part1_output <- furrr::future_map(.x = a$row, .f = part1_fun, .progress = TRUE) | |
| part1_output <- rbindlist(part1_output) | |
| # part1_output <- pblapply(X= a$row, FUN = part1_fun) | |
| head(part1_output) | |
| # merge | |
| tt[part1_output, on=c('fromId', 'travel_time'), pij_part1 := i.pij_part1 ] | |
| tt[, pij_part1 := fifelse(is.na(pij_part1), 0, pij_part1)] | |
| head(tt) | |
| # part 2 | |
| tt[, pij_part2 := pij_part1 - pop_dest, by=.(fromId, toId)] | |
| ### calculate radiation model -------------------------------------------------------- | |
| tt[, rad := Ti * ((pop_orig * pop_dest) / ((pop_orig + pij_part2) * (pop_orig + pop_dest + pij_part2))), by=.(fromId, toId)] | |
| summary(tt$rad) | |
| ### check results -------------------------------------------------------- | |
| # remove Inf values | |
| df <- subset(tt, rad < Inf) | |
| summary(df$rad) | |
| summary(df$travel_time) | |
| plot( density(df$travel_time) ) | |
| plot( density(df$rad) ) | |
| # plot a sample of the results | |
| sample_size <- 0.10 | |
| draw <- sample(x = 1:nrow(df), size = sample_size * nrow(df), replace = F) | |
| df2 <- df[draw,] | |
| ggplot() + | |
| geom_density(data=df2, aes(x=travel_time, weight=rad), alpha=.1) | |
| ggplot() + | |
| geom_pointdensity(data=df2, aes(x=travel_time , y=rad), alpha=.1) + | |
| scale_color_viridis_c() + | |
| scale_y_log10() |
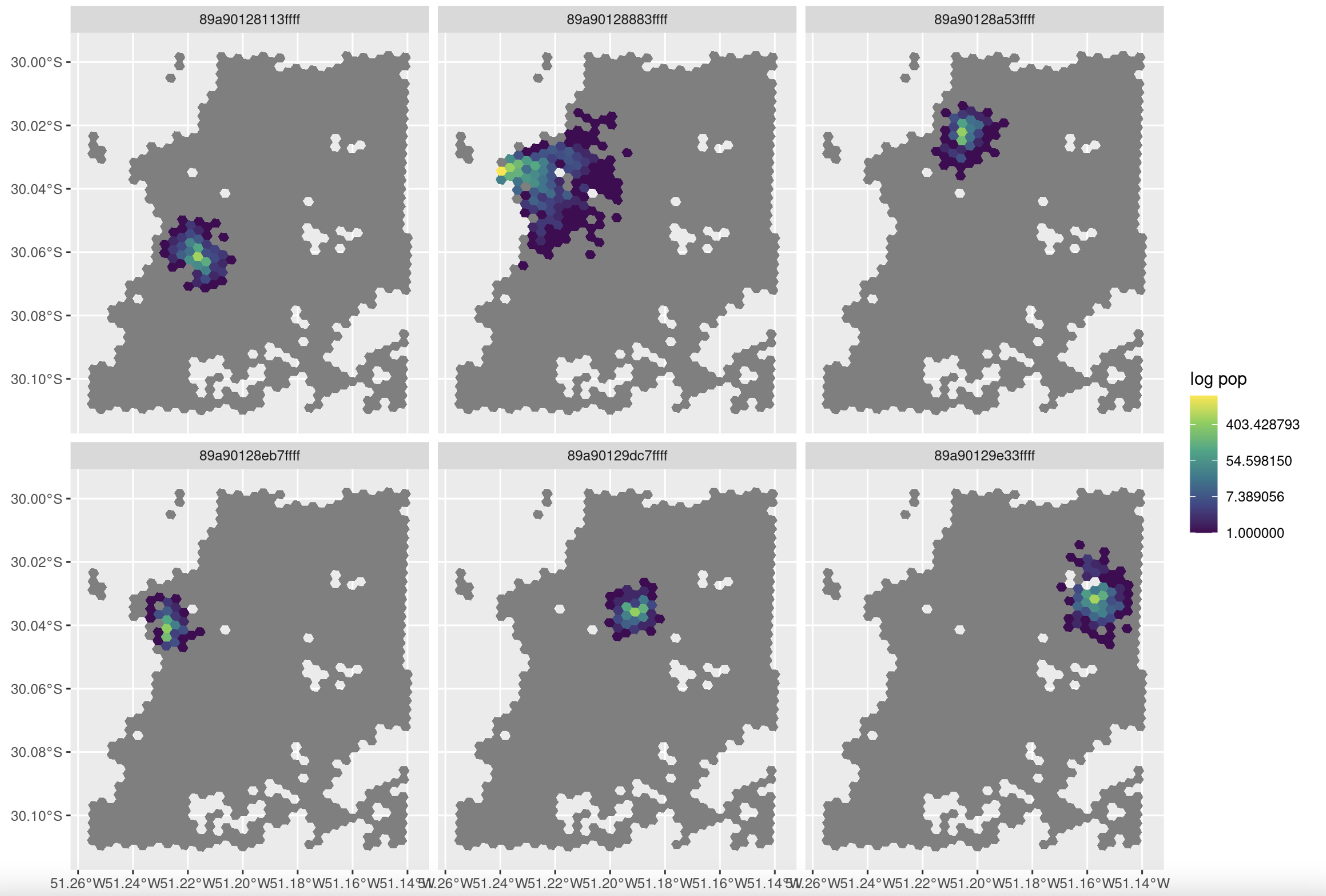
I've mapped some flows, and it looks like people are not very interested in working far from home:
Code:
# Map results
library(h3jsr)
library(sf)
tt[, rad_pop := round(rad * pop_total)]
p_hex <- h3jsr::h3_to_polygon(points$id, simple = FALSE)
s_origins = sample(points$id, size = 6, prob = points$population)
df3 <- tt[fromId %in% s_origins & rad_pop > 0]
df3$geometry <- h3jsr::h3_to_polygon(df3$toId)
df3 <- st_as_sf(df3, crs = 4326)
ggplot(df3) +
geom_sf(data = p_hex, fill = "grey50", color = NA) +
geom_sf(aes(fill = rad_pop), color = NA) +
facet_wrap(~fromId) +
scale_fill_viridis_c(trans = "log") +
labs(fill = "log pop")
thanks, Marcus! I've updated the code to incorporate the corrections you suggested !
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment
Uh oh!
There was an error while loading. Please reload this page.