-
-
Save rougier/e5eafc276a4e54f516ed5559df4242c0 to your computer and use it in GitHub Desktop.
| # ----------------------------------------------------------------------------- | |
| # From https://en.wikipedia.org/wiki/Minkowski–Bouligand_dimension: | |
| # | |
| # In fractal geometry, the Minkowski–Bouligand dimension, also known as | |
| # Minkowski dimension or box-counting dimension, is a way of determining the | |
| # fractal dimension of a set S in a Euclidean space Rn, or more generally in a | |
| # metric space (X, d). | |
| # ----------------------------------------------------------------------------- | |
| import scipy.misc | |
| import numpy as np | |
| def fractal_dimension(Z, threshold=0.9): | |
| # Only for 2d image | |
| assert(len(Z.shape) == 2) | |
| # From https://github.com/rougier/numpy-100 (#87) | |
| def boxcount(Z, k): | |
| S = np.add.reduceat( | |
| np.add.reduceat(Z, np.arange(0, Z.shape[0], k), axis=0), | |
| np.arange(0, Z.shape[1], k), axis=1) | |
| # We count non-empty (0) and non-full boxes (k*k) | |
| return len(np.where((S > 0) & (S < k*k))[0]) | |
| # Transform Z into a binary array | |
| Z = (Z < threshold) | |
| # Minimal dimension of image | |
| p = min(Z.shape) | |
| # Greatest power of 2 less than or equal to p | |
| n = 2**np.floor(np.log(p)/np.log(2)) | |
| # Extract the exponent | |
| n = int(np.log(n)/np.log(2)) | |
| # Build successive box sizes (from 2**n down to 2**1) | |
| sizes = 2**np.arange(n, 1, -1) | |
| # Actual box counting with decreasing size | |
| counts = [] | |
| for size in sizes: | |
| counts.append(boxcount(Z, size)) | |
| # Fit the successive log(sizes) with log (counts) | |
| coeffs = np.polyfit(np.log(sizes), np.log(counts), 1) | |
| return -coeffs[0] | |
| I = scipy.misc.imread("sierpinski.png")/256.0 | |
| print("Minkowski–Bouligand dimension (computed): ", fractal_dimension(I)) | |
| print("Haussdorf dimension (theoretical): ", (np.log(3)/np.log(2))) |
Hi there,
I would like to know that what is the use of threshold=0.9 value, why we are using this value.
Better to use imageio.imread in line 52 as imread is deprecated in SciPy 1.0.0.
Does this code work? I have tried running it on a 2D box generated as follows:
seg = np.zeros(shape = (100,100))
seg[20:80,20:80] = 1.0
The function returned 'nan'. I was expecting something close to 2.0.
When I ran it on an irregular segmentation, I got fractal dimensions between 0.45 - 0.9. The reference values I found in the literature for the same type of segmentation are around 1.2.
Do you have the log (reason for NaN)?
When I run this:
`seg = np.zeros(shape = (100,100))
seg[20:80,20:80] = 1.0
fractal_dimension(seg)`
In a Jupyter notebook I get
"RuntimeWarning: divide by zero encountered in log
plt.plot(np.log(sizes), np.log(counts))"
It seems there is a problem with trying to fit nearly linear data.
Hi,
I looked at this a bit more today. the problem is one of the "counts" is 0 so log(counts) = -inf :
counts =
[1.38629436 2.7080502 2.99573227 3.68887945 -inf]
I can remove the -inf term using this:
x = np.log(sizes)
y = np.log(counts)
mask = ~np.isinf(y)
x = x[mask]
y = y[mask]
coeffs = np.polyfit(x, y, 1)
return -coeffs[0]
The result is I get D = 1.038. I was expecting it to be closer to 2.
Yes, I would have expected 2. Do you have by any change a reference implementation somewhere such that we can check?
Here is the code I'm using :
https://repl.it/repls/AlarmingHarmlessScience
Thank you for your help with this!!
Looking at https://en.wikipedia.org/wiki/Minkowski%E2%80%93Bouligand_dimension, I think I may have computer the dimension of the border and not the actual shape because I filtered out the full boxes. This may explain why the result is 1.
If I run on a rectangle, I get something around 0.55. I think it should be pretty close to either 1 or 2 depending how you did it. To be honest I haven't had time to review your code in depth. I am actually analyzing a 3D shape and using fractal dimension as a measure of surface irregularity, so I'm using a 3D fractal dimension code I found elsewhere for that. So this is not a high priority for me. However I thought it might be worth looking at the fractal dimension for 2D slices of the object as well.
I am getting this error message when executing this programme in PyCharm.
1)"AttributeError: module 'scipy.misc' has no attribute 'imread'"
2) "AttributeError: module 'scipy' has no attribute 'misc'"
@akshahi17 Probably the imread function has been deprecated in scipy. You cn now use the imageio library instead.
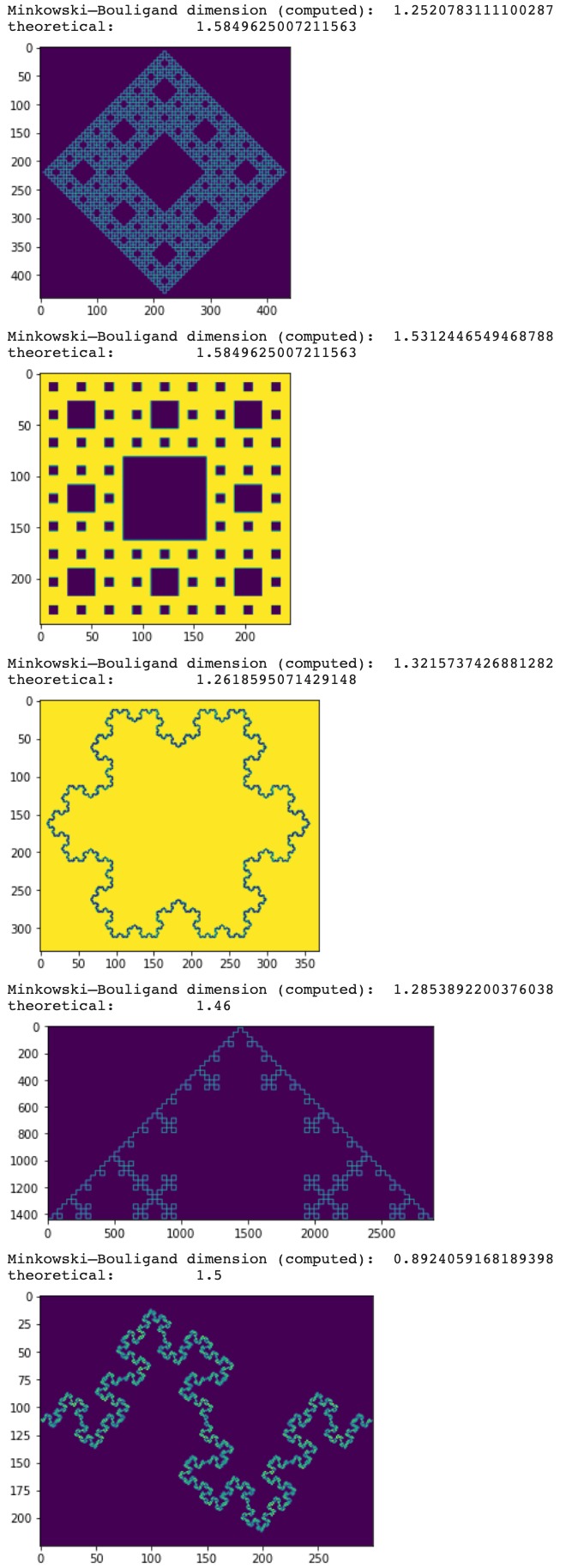
Hi, I was testing it on some fractals with defined dimensions, below is my code. The problem is that it's far from the dimensions and even for the same fractal in a different rotation (Sherpinski), it is not giving the same values (which I believe should, this should be independent of rotation). Where is the problem?
Also, I believe that it should go to squares 2x2, that is:
sizes = 2**np.arange(n, 2, -1) should become sizes = 2**np.arange(n, 0, -1).
Source:
https://en.wikipedia.org/wiki/Sierpi%C5%84ski_carpet
https://en.wikipedia.org/wiki/Koch_snowflake
# -----------------------------------------------------------------------------
# From https://en.wikipedia.org/wiki/Minkowski–Bouligand_dimension:
#
# In fractal geometry, the Minkowski–Bouligand dimension, also known as
# Minkowski dimension or box-counting dimension, is a way of determining the
# fractal dimension of a set S in a Euclidean space Rn, or more generally in a
# metric space (X, d).
# -----------------------------------------------------------------------------
import numpy as np
import matplotlib.pyplot as plt
def fractal_dimension(Z, threshold=0.9):
# Only for 2d image
assert(len(Z.shape) == 2)
# From https://github.com/rougier/numpy-100 (#87)
def boxcount(Z, k):
S = np.add.reduceat(
np.add.reduceat(Z, np.arange(0, Z.shape[0], k), axis=0),
np.arange(0, Z.shape[1], k), axis=1)
# We count non-empty (0) and non-full boxes (k*k)
return len(np.where((S > 0) & (S < k*k))[0])
# Transform Z into a binary array
Z = (Z < threshold)
# Minimal dimension of image
p = min(Z.shape)
# Greatest power of 2 less than or equal to p
n = 2**np.floor(np.log(p)/np.log(2))
# Extract the exponent
n = int(np.log(n)/np.log(2))
# Build successive box sizes (from 2**n down to 2**1)
sizes = 2**np.arange(n, 1, -1)
# Actual box counting with decreasing size
counts = []
for size in sizes:
counts.append(boxcount(Z, size))
# Fit the successive log(sizes) with log (counts)
coeffs = np.polyfit(np.log(sizes), np.log(counts), 1)
return -coeffs[0]
special = [
'https://upload.wikimedia.org/wikipedia/commons/thumb/7/7c/Peano_Sierpinski_carpet_4.svg/440px-Peano_Sierpinski_carpet_4.svg.png',
'https://upload.wikimedia.org/wikipedia/commons/thumb/3/3b/Quadratic_Koch_2.svg/2880px-Quadratic_Koch_2.svg.png',
'https://upload.wikimedia.org/wikipedia/commons/thumb/5/55/Quadratic_Koch.svg/300px-Quadratic_Koch.svg.png'
]
images = [
'https://upload.wikimedia.org/wikipedia/commons/thumb/7/7c/Peano_Sierpinski_carpet_4.svg/440px-Peano_Sierpinski_carpet_4.svg.png',
'https://upload.wikimedia.org/wikipedia/commons/thumb/b/b7/Sierpinski_carpet_3.svg/244px-Sierpinski_carpet_3.svg.png',
'https://upload.wikimedia.org/wikipedia/commons/f/f0/Flocke.PNG',
'https://upload.wikimedia.org/wikipedia/commons/thumb/3/3b/Quadratic_Koch_2.svg/2880px-Quadratic_Koch_2.svg.png',
'https://upload.wikimedia.org/wikipedia/commons/thumb/5/55/Quadratic_Koch.svg/300px-Quadratic_Koch.svg.png'
]
dimensions = [(np.log(3)/np.log(2)), (np.log(3)/np.log(2)), (np.log(4)/np.log(3)), 1.46, 1.5]
for dim, im in zip(dimensions, images):
# load im
x = plt.imread(im)
if im in special:
print("Minkowski–Bouligand dimension (computed): ", fractal_dimension(x[:,:,3]))
print("theoretical: ", dim)
plt.imshow(x[:,:,3])
plt.show()
else:
print("Minkowski–Bouligand dimension (computed): ", fractal_dimension(x[:,:,1]))
print("theoretical: ", dim)
plt.imshow(x[:,:,1])
plt.show()and the result:
Minkowski–Bouligand dimension (computed): 1.2520783111100287
theoretical: 1.5849625007211563
Minkowski–Bouligand dimension (computed): 1.5312446549468788
theoretical: 1.5849625007211563
Minkowski–Bouligand dimension (computed): 1.3215737426881282
theoretical: 1.2618595071429148
Minkowski–Bouligand dimension (computed): 1.2853892200376038
theoretical: 1.46
Minkowski–Bouligand dimension (computed): 0.8924059168189398
theoretical: 1.5
I'm not sure but the images seems to be a bit noisy which might impact the computation. Can you try to make black and white (for example I = I > 0.5 and check if the result is the same? If this is the case, this means we need some threshold value when deciding if a box is empty or not.
Except for the first and last, they are all binary (ie B&W). I have tried and you were right, @rougier, about those, setting the threshold to 0.2 works on them.
What about this note?
Also, I believe that it should go to squares 2x2, that is:
sizes = 2**np.arange(n, 2, -1)should becomesizes = 2**np.arange(n, 0, -1).
I am curious because you used to have it that way (at least that is what I see in the history) but you have a discrepancy in the note and in the code. Your code says:
2**np.arange(n, 1, -1) # eg for n=4 you get array([16, 8, 4])but the comment says
Build successive box sizes (from 2**n down to 2**1) which suggest this code:
2**np.arange(n, 0, -1) # eg for n=4 you get array([16, 8, 4, 2])And that I believe is the correct code.
Code
import numpy as np
import matplotlib.pyplot as plt
def fractal_dimension(Z, threshold=0.9):
# Only for 2d image
assert(len(Z.shape) == 2)
# From https://github.com/rougier/numpy-100 (#87)
def boxcount(Z, k):
S = np.add.reduceat(
np.add.reduceat(Z, np.arange(0, Z.shape[0], k), axis=0),
np.arange(0, Z.shape[1], k), axis=1)
# We count non-empty (0) and non-full boxes (k*k)
return len(np.where((S > 0) & (S < k*k))[0])
# Transform Z into a binary array
Z = (Z < threshold)
plt.imshow(Z)
plt.show()
# Minimal dimension of image
p = min(Z.shape)
# Greatest power of 2 less than or equal to p
n = 2**np.floor(np.log(p)/np.log(2))
# Extract the exponent
n = int(np.log(n)/np.log(2))
# Build successive box sizes (from 2**n down to 2**1)
sizes = 2**np.arange(n, 1, -1)
# Actual box counting with decreasing size
counts = []
for size in sizes:
counts.append(boxcount(Z, size))
# Fit the successive log(sizes) with log (counts)
coeffs = np.polyfit(np.log(sizes), np.log(counts), 1)
return -coeffs[0]
special = [
'https://upload.wikimedia.org/wikipedia/commons/thumb/7/7c/Peano_Sierpinski_carpet_4.svg/440px-Peano_Sierpinski_carpet_4.svg.png',
'https://upload.wikimedia.org/wikipedia/commons/thumb/3/3b/Quadratic_Koch_2.svg/2880px-Quadratic_Koch_2.svg.png',
'https://upload.wikimedia.org/wikipedia/commons/thumb/5/55/Quadratic_Koch.svg/300px-Quadratic_Koch.svg.png'
]
images = [
'https://upload.wikimedia.org/wikipedia/commons/thumb/7/7c/Peano_Sierpinski_carpet_4.svg/440px-Peano_Sierpinski_carpet_4.svg.png',
'https://upload.wikimedia.org/wikipedia/commons/thumb/b/b7/Sierpinski_carpet_3.svg/244px-Sierpinski_carpet_3.svg.png',
'https://upload.wikimedia.org/wikipedia/commons/f/f0/Flocke.PNG',
'https://upload.wikimedia.org/wikipedia/commons/thumb/3/3b/Quadratic_Koch_2.svg/2880px-Quadratic_Koch_2.svg.png',
'https://upload.wikimedia.org/wikipedia/commons/thumb/5/55/Quadratic_Koch.svg/300px-Quadratic_Koch.svg.png'
]
dimensions = [(np.log(3)/np.log(2)), (np.log(3)/np.log(2)), (np.log(4)/np.log(3)), 1.46, 1.5]
for dim, im in zip(dimensions, images):
# load im
x = plt.imread(im)
if im in special:
print("Minkowski–Bouligand dimension (computed): ", fractal_dimension(x[:,:,3], threshold=0.2))
print("theoretical: ", dim)
print(f"min {x[:,:,3].min()} and max {x[:,:,3].max()} in the array/image")
else:
print("Minkowski–Bouligand dimension (computed): ", fractal_dimension(x[:,:,1],threshold=0.2))
print("theoretical: ", dim)
print(f"min {x[:,:,1].min()} and max {x[:,:,1].max()} in the array/image")I am getting this error message when executing this programme in PyCharm.
1)"AttributeError: module 'scipy.misc' has no attribute 'imread'"
2) "AttributeError: module 'scipy' has no attribute 'misc'"
@akshahi17 check this answer https://stackoverflow.com/questions/15345790/scipy-misc-module-has-no-attribute-imread
the function has been deprecated so you can either downgrade (not recommended) or use what @rougier recommended (imageio) or what I'm using, imread in matplotlib.pyplot.
Hi! I found this code very useful and I would like to cite it in my paper. Is there a correct way to cite this? Please help! I have to submit soon!
Glad the code is useful but I'm afraid there there's no proper way to cite a it. Maybe as a misc bibitem with the relevant url/author.
hello
sorry, can anyone tell me how to change the data that I want to get from the link to my laptop data?
I want to try to extract features of an iris
thanks, @jankaWIS
and sorry if I ask again is the 52 line is
images = [
'https://upload.wikimedia.org/wikipedia/commons/thumb/7/7c/Peano_Sierpinski_carpet_4.svg/440px-Peano_Sierpinski_carpet_4.svg.png',
'https://upload.wikimedia.org/wikipedia/commons/thumb/b/b7/Sierpinski_carpet_3.svg/244px-Sierpinski_carpet_3.svg.png',
'https://upload.wikimedia.org/wikipedia/commons/f/f0/Flocke.PNG',
'https://upload.wikimedia.org/wikipedia/commons/thumb/3/3b/Quadratic_Koch_2.svg/2880px-Quadratic_Koch_2.svg.png',
'https://upload.wikimedia.org/wikipedia/commons/thumb/5/55/Quadratic_Koch.svg/300px-Quadratic_Koch.svg.png'
]
?
sorry I using the jupyter notebook so not to know about the number
Why are boxes of 2x2 excluded?
Hello, thank you very much for this function. I am finding it very useful in my study of fractals.
However, I drew a jagged line and tried to compute the fractal dimension
Using this code, I get a dimension of around 0.94 which is clearly wrong as it is < 1.
Why am I getting such a number?
Additionally, with other free fractal analysis software, I get a dimension of around 1.088
FWIW the calculation is approximate.. also look at my comment above, there might still be a subtle issue with this code.
Hi delton137!
I used another simple line that I generated, and then with altering the number of boxes, and then after iterating and computing the mean, I believe I got a better estimate. For this other curve, the box-count dimension was 1.06 ish. And it was well within the 95% CI range when I compared it to the FD from the one measured by the software.
I think I hopefully solved my own problem. I came across some papers re: optimal sizes of boxes. I will need to see how to implement that!
Also, I saw that you are also working in medical imaging. It is very inspiring to see interest in fractals and medical imaging. I am but a mere medical student currently, I hope to see a lot more fractals in the future, and a lot more collaboration!


Hi, this piece of code is really useful.
Please can you help me out, I am having trouble in figuring out the "boxcount" method written.
My particular confusion lies in the return statement, "return len(np.where((S > 0) & (S < kk))[0])" S>0 implies that there is some value in that pixel, but what about the later part i.e. S<kk[0].
Thank you in advance :)